Research Article - Imaging in Medicine (2017) Volume 9, Issue 4
Breast Cancer Heterogeneity Analysis as Index of Response to Treatment Using MRI Images: A Review
Mohammed El Adoui1*, Stylianos Drisis2, Mohamed Amine Larhmam1, Marc Lemort2 & Mohammed Benjelloun11Computer Science Unit, Faculty of Engineering, University of Mons, Belgium
2Radiology Department, Jules Bordet Institute université libre de Bruxelles, Belgium
- Corresponding Author:
- Mohammed El Adoui
Computer Science Unit
Faculty of Engineering, University of Mons, Belgium
E-mail: mohammed.eladoui@umons.ac.be
Abstract
Purpose: Breast tumor structure contains a high degree of heterogeneity. This heterogeneity has been correlated with the level of tumor response to neoadjuvant chemotherapy. A significant number of studies using magnetic resonance imaging have looked into the quantification of intra tumor heterogeneity in breast cancer. Nevertheless, a limited number of these studies have specifi-cally looked at evaluating breast cancer heterogeneity as a biomarker of response to treatment.
Methods: In this paper, several heterogeneity quantification methods, which have studied breast cancer response to treatment through MR images, will be presented. Important methodological and technological techniques using experimental design, such as histogram analysis, texture analysis and parametric response mapping (PRM), will be explored.
Results: Data acquisition, the number of patients, the number of treatment cycles, and other analysis will be discussed for each presented research case. Furthermore, some proposed methods will be evaluated on our institution MRI dataset, collected between 2013 and 2016.
Conclusion: This paper can be used as a guideline for investigators working on breast cancer het-erogeneity as a biomarker of response to treatment.
Keywords
breast cancer imaging ▪ MRI ▪ intratumor heterogeneity ▪ texture analysis ▪ histogram analysis ▪ PRM
Introduction
Intra-tumor heterogeneity analysis presents a great challenge for the characterization of breast cancer’s response to treatment [1-3]. Several techniques have been documented to identify intra-tumor genetic heterogeneity or temporal diversity for breast tumor cells [4,5]. There is also evidence to demonstrate that, in breast cancers, sub-clonal cancerous cells may exist in different regions of a tumor (spatial heterogeneity), or may appear after one or many cycles of chemotherapy (temporal heterogeneity) [6,7]. However, while imaging is essential to diagnosis, staging and response assessment, most clinical radiology, and research studies only measure tumor size by response evaluation criteria in relation to solid tumor response (RECIST) [8]. Neoadjuvant chemotherapy (NAC) is commonly recommended for advanced breast cancers, with the aim of facilitating conservative surgery and achieving more survival [9,10]. Magnetic Resonance Imaging (MRI) technique produces a variety of image types, such as T1-weighted, T2-weighted, dynamic contrast enhanced and diffusion weighted images [11-15]. Each of these sequences is relevant to quantifying breast tumor heterogeneity. Therefore, several studies have shown that MRI has better diagnostic accuracy than other clinical breast examinations, such as ultrasonography and mammography [16-20]. Indeed, functional imaging sequences of MRI enable a greater lesion to background enhancement.
Breast cancer heterogeneity has been quantified using several image analysis techniques: 1-histogram analysis is used to compute many features, such as standard deviation (SD) and percentile values; 2-texture analysis is used to quantify the breast tumor heterogeneity by studying the spatial variation of tumors; 3-parametric response mapping is used to compare the baseline and post-treatment examinations [21-23]. In this paper, an overview of works concerned with the study of MRI breast tumor heterogeneity as a biomarker of response to neoadjuvant treatment will be presented. For each study, the patient number, MRI timing and acquisition, reference, and the results found are presented in some comprehensive tables.
Methods
While there are several methods used for breast imaging in the literature, here the focus is on the use of MR images for intra tumor heterogeneity as index of breast cancer assessment. In this review paper, we included only publications related to the breast tumor heterogeneity assessment using MR images to predict the response to treatment using images from a considerable number of patients as data. The studies have been classified into three categories based on the heterogeneity quantification techniques used: Histogram Analysis (HA), Texture Analysis (TA) and Parametric Response Mapping (PRM).
■ Histogram analysis
Histogram analysis describes the statistical information of an image or a region/volume of interest. Indeed, intra-tumor heterogeneity can be quantified by analyzing histogram shape of the volume of interest (VOI). Most histogram analysis technique uses descriptive parameters to characterize and compare distributions of tumor voxels. This analysis provides quantitative measures, such as standard deviation, kurtosis, and skewness. These parameters represent the first-order statistical properties of images [24].
Many studies have concluded that histogram analysis contributes to discriminating between benign and malignant tumors [25,26]. However, few of them correlate histogram based measures, such as kurtosis, skewness and percentile, with a response to treatment [27].
In this context, Johansen et al. [28] performed a retrospective study including 24 patients with breast cancer, scanned before and after the first cycle of chemotherapy, using data from dynamic contract enhanced MR images (DCE-MRI). Images were acquired 3 to 5 min after the injection of the contrast agent. Relative signal intensity (RSI) histograms and AUC were calculated from the DCE-MRI curve, from which five measures were extracted (mean, standard deviation, skew and kurtosis) and used for 5 year survival prediction. Clinical response (CR) was used as a reference, and it was concluded that skewness and kurtosis are strongly correlated to the response to treatment by using KNN and PNN (Kohonen and probabilistic neural network) methods. The sensitivity and specificity of this approach were between 80% and 92%, respectively. However, in this study the spatial relationship between voxels was ignored by the histogram analysis approach.
Padhani et al. [29] performed a retrospective study of 25 patients with primary breast cancer before and after the first cycle of treatment. The T1-weighted sequence acquired at 2 min and 42 s after the injection of the contrast. Treatment response in breast cancer patients was more sensitive using the range of the Ktrans [30] histogram, compared with the use of the median of Ktrans. Receivers operating characteristic (ROC) analysis of the tumor size were used to identify patients who showed no response to chemotherapy. It was demonstrated that responsive patients showed a decrease in the range of Ktrans histogram values. Furthermore, non-responsive patients showed a widening of the Ktrans histogram.
Hayes et al. [31] used DCE-MRI sequence on 15 patients with breast cancer before and after one chemotherapy cycle. In this study, regions of interest (ROIs) were delineated on the subtracted images. Changes in the temporal pattern of signal enhancement, the amplitude of enhancement, and Ktrans were determined. Ktrans histogram was generated using the whole tumor ROI before and after treatment. This histogram was used to evaluate tumor heterogeneity. A decrease in the high Ktrans values in responsive patients after one cycle of treatment, and an increase in the number of lower Ktrans were observed. Therefore, in non-responsive patients, the opposite effect was observed, with a shift in histogram distribution to the right (higher Ktrans values). It was concluded that histogram distribution features other than the median value can provide additional information regarding the mechanism of tumor response. A greater range of changes in the absolute value of Ktrans was observed in the upper extremes of the histogram (‘hot-spots’) compared with the median values.
The tumor nature represents spatial heterogeneity. However, the above studies showed results based only on the histogram technique, which ignores the spatial relationship between the tumor voxels. Indeed, an identical histogram can present two different tumors, for example, a tumor with identical sub-regions could have a similar histogram with a tumor having many dispersed features over the integral mass. Therefore, spatial distribution information is recommended to differentiate this type of lesion [32-34].
■ Texture analysis
In this section, an overview of research studies using texture analysis approach to quantify breast cancer response in MR images [35-42] will be presented.
Texture analysis methods are used to study the distribution of individual voxels within a tumor region and provide their spatial relationship. Texture analysis in MRI data is frequently based on the Gray-Level Co-occurrence Matrix (GLCM). The latter was first described by Haralick et al. [33]. In this method, a matrix is defined over an image (or ROI) to provide the distribution of co-occurring pixel/voxel values, which provides information about their spatial relationship. Many textural features, such as entropy, correlation, uniformity, variance, and homogeneity and 14 other features, can be extracted from the GLCM [34].
In this context, Golden et al. [35] performed a retrospective study to predict four types of breast cancer response in 60 patients using a dynamic protocol of DCE-MRI. These patients were receiving NAC treatment. Quantitative lesion analysis was achieved by extracting four GLCM texture features from six kinetic maps of each tumor volume. The reference of treatment was determined on the pathologic complete response (pCR) as 22.37% and 25.83% for residual invasive tumor and/or positive lymph node. The authors concluded that the heterogeneity in kinetic maps could reflect heterogeneity in the physiology of the tumors.
This study was performed using twodimensional images instead of three dimensional ones. The number of cycles was not the same for every patient, in fact among the 60 patients there were 12 patients who were treated with 4 cycles and the 48 remaining patients underwent treatment for 6 cycles of treatment.
Teruel et al. [36] studied the potential of texture analysis using DCE-MRI data to predict clinical and pathological responses to NAC after 4 treatment cycles. A retrospective study of 58 patients with advanced breast cancer was performed. Each lesion was segmented semi-automatically using 2 min post-contrast subtracted images. Using four GLCMs, 16 texture features were determined over tumor volumes at each non-subtracted post-contrast time point. Firstly, for each slice containing tumor, histogram equalization was applied, accounting for the segmented tumor region. Secondly, four 32*32 GLCMs were calculated for each image. Prediction of the clinical and pathological response was based on multiple statistical tests (q-values and p-values). Significant results were found at 3 min postcontrast for various texture features. Four texture features (sum variance, sum entropy, entropy and difference variance) were found to be significant at the 2 min time point, with an area under the curve (AUC) of 0.69.
Michoux et al. [37] performed a retrospective study of 69 cases with invasive ductal carcinoma of the breast receiving pre-treatment chemotherapy using DCE-MRI. One pre- and five post-injection images were acquired with a temporal resolution of 1 min after injection of the contrast agent. The total acquisition time for the protocol was about 6 min. Pathological complete response (pCR) was defined by the absence of invasive cancer in breast and nodes. DCE-MRI was performed at 1.5T with a fatsuppressed sequence. Visual texture (GLCM and Run Length Matrix (RLM) [38]), BI-RADS and kinetic parameters were measured in each region of interest. The prediction model, based on the 4 parameters (inverse difference moment, Gray-Level Non-uniformity, Long Run High Gray-Level Emphasis, wash-in) and k-means clustering as a statistical classifier, identified non-responsive patients with a high sensitivity of 84%. Biological markers and histological grades, combined with texture and kinetic parameters, did not yield an improvement of the prediction accuracy. However, the authors point out that the biological parameters should not be ignored.
Afrane et al. [39] performed a retrospective study of a quantitative T1-weighted DCE-MRI for 100 patients with breast cancer, to predict the response to treatment through texture analysis. Texture analysis was performed on pre-contrast and on 1 to 5 min's post-contrast images. Software was developed to use texture features based on GLCM. Patients were classified in terms of their level of response to treatment: partial responders (PR) corresponding to a decrease in tumor size of 50% and non-responders (NR) correspond to a decrease of less than 50%. ROIs were generated semi-automatically on all slices from DCE-MRI sequences. For texture analysis, ROIs from breast images were used on several slices, to provide an assessment of the whole tumor volume.
Minkowski functionals [40] is another texture analysis technique which can be an alternative to GLCM. In this method, instead of analyzing the MR images directly, a series of binary images are used. These images are extracted from MR scans using a rising threshold to remove irrelevant pixels. Minkowski Functionals enables to extract only three texture features, compared to the GLCM method which can provides more than 16 features.
Micheal et al. [40] performed a retrospective study, using texture analysis of DCE-MRI data from 100 patients with breast cancer. A quantitative fat-nulled T1-weighted sequence was used in this study. Images taken from the 2nd or 3rd post-contrast phase were acquired 1 min after injection of the contrast agent. Instead of using classic GLCM for each binary image, three Minkowski Functionals values (area, perimeter, euler value) were calculated. Indeed, the change of binary threshold was raised using 6th order polynomials. These polynomials provided the five patients’ subgroups: Triple- Negative Breast Cancer (TNBC) status, chemotherapy response, biopsy grade, nodal status, and lymph vascular invasion status. It was confirmed that the Minkowski Functionals method provided several significant differences in the TNBC, biopsy grade and lymph vascular invasion status, with a p-value lower than 0.05.
Parikh et al. [41] performed a retrospective study using texture features derived from DCE-MRI data of 36 patients with primary breast cancer. These patients were receiving neoadjuvant chemotherapy (NACT). T2 and T1-weighed DCE-MRI imaging was performed before treatment, mid-treatment (after three NACT cycles) and after treatment. The images were acquired 3 min after the injection of the contrast agent. Changes in MRI imaging heterogeneity were correlated with pathologic complete responders (pCR) and compared with the standard method (RECIST). Changes in tumor entropy (irregularity) and uniformity (gray-level distribution) were determined before and after MRI image filtration. Entropy and uniformity for pCR and non-responders were compared using ROC curve. Images of all patients were filtered to highlight different size characteristics (from 2 to 12 pixels). Treatment results show an increase in uniformity and a decrease in entropy in all images, using several filters. In fact, the results of uniformity and entropy were as flow: uniformity: 23.45% and 22.62%; entropy: 219.15% and 219.26%.
Jae-Hun et al. [42] performed a retrospective study using texture analysis of the entire primary breast tumor in 203 patients. T2-weighted and contrast-enhanced T1-weighted subtraction sequences, acquired 90 s after contrast injection, were used. MR imaging was performed with a 1.5 T. To quantify tumor heterogeneity, uniformity and entropy features were calculated from T2- weighted, contrast-enhanced and T1-weighted subtraction images for all ROIs. Indeed, entropy is a measure that can reflect tumor texture irregularity, and uniformity. Entropy can also show how gray levels are distributed in an image. A 10-fold cross-validation [36] was used on the image dataset to avoid over-fitting and determine cut-off points in the ROC curve.
■ Parametric response mapping (prm)
Parametric Response Mapping (PRM) is a new method which enables the evaluation of a patient’s response to chemotherapy. PRM measures changes of quantitative MRI values in underlying tumor tissues by using a voxel-byvoxel approach. This method is mainly intended to compare pre- and post-treatment VOI. This comparison is performed using image coregistration technique, at short time intervals after the initiation of treatment, to provide not only an early assessment of the treatment outcome, but also information of intra-tumor heterogeneity [43]. This technique has not been widely used for breast cancer. However, the technique has proven its efficiency in brain cancer research.
Cho et al. [44] presented a prospective study of 48 patients with breast cancer was performed. T1-weighted sequence (7 min after contrast agent injection) was used before and after the first cycle of NACT. The pathological response was observed after surgery. The tumor size was measured on DCE-MRI images, and was considered as the same tumor size after the chemotherapy. Non-linear automatic registration was performed for each volume obtained before treatment (baseline) and after the first cycle of chemotherapy. Co-registration technique was evaluated and validated, first, by using the toggle view, and second, by blended volume visualization or checkerboard view tools. PRM analysis was performed by calculating the interval change between the baseline and the first cycle signal intensity, using a voxel-to-voxel comparison. The results show that among the 48 patients evaluated, 6 patients achieved pCR and 42 were non-pathological responders (npCR). 38 patients showed a good response, with a grade above 3, and 10 patients demonstrated a low response, with a grade of 1 or 2. PRM prediction of good response showed an AUC of 0.72. One of limitation of this method is the high processing time due to manual segmentation and co-registration of images.
Boes et al. [45] presented a new approach for the co-registration of images pre- and post-treatment using MRI diffusion-weighted imaging (DWI) was presented. A deformable coregistration was based on the thin-plate splines (TPSs) geometric technique. The data was obtained from several multi-center prospective clinical trials including 52 patients, adjusted for 4 to 5 min scans. This approach can be used to develop and validate PRM-based methods as early response metrics for the prediction of breast cancer responders from non-responders using DWI and DCE-MRI images.
Results
In this section, three comparative tables of the methods discussed in the previous sections are presented: TABLE 1 (Histogram Analysis), TABLE 2 (Texture Analysis) and TABLE 3 (PRM). For each method, the number of patients, MRI timing, technical details and findings are presented. Of these studies, 25% reported the use of Histogram Analysis, 58,33% Texture Analysis and 16,67% PRM.
| Author | Patients | Timing | Method | Acquisition | Study | Reference | Findings |
|---|---|---|---|---|---|---|---|
| Johansen et al. [28] | 24 | 1 Cycle | Histogram Analysis of RSI of DCE-MRI (semi-automated segmentation) |
DCE-MRI-T1 (3-5 min after injection of contrast agent) |
Retro | CR | - Reduction of AUC after 1 cycle of chemotherapy |
| Padhani et al. [29] | 25 | 1-2 Cycles | Histogram Analysis of Ktrans DCE-MRI values (semi-automated segmentation) |
DCE-MRI-T1 (2 min 42 s after injection of contrast agent) |
Retro | pCR | - Absence of pCR was improved by tumor size and Ktrans values - AUC for size: 0.93 - AUC for Ktrans Range: 0.94 |
| Hayes et al. [31] | 15 | 1 Cycle | Histogram Analysis of pixel maps of K(trans) (semi-automated segmentation) |
DCE-MRI-T1 (1 min after injection of contrast agent) |
Retro1 | pCR | - Changes in the rate of enhancement and Ktrans values were correlated by more than 50%. - Negative correlation between Ktrans values and response to treatment. |
1Retrospective/Prospective
2Cycle of chemotherapy
3Relative signal intensity
4Clinical Response
5Area under the curve
6Pathological Complete Response
7Volume transfer coefficient reflecting vascular permeability
Table 1: Literature on the histogram analysis technique for assessing breast cancer response.
| Author | Patients | Timing | Method | Acquisition | Study | Reference | Finding |
|---|---|---|---|---|---|---|---|
| Golden et al. [34] | 60 | 4-6 Cycles | GLCM (Semi-Automated segmentation) |
DCE-MRI-T2 (2 min 42 s after injection of contrast agent) |
Retro | pCR+Lymph Nodes | - pCR was determined as reference to treatment - Texture features extracted after chemotherapy predicted a residual invasive tumor - Correlation between Kinetic maps and tumor physiology heterogeneities. |
| Teruel et al. [35] | 58 | 4 Cycles | GLCM (Semi-Automated segmentation) |
DCE-MRI-T1 (2 min after injection of contrast agent) |
Retro | pCR+cR | - 8 texture features were significant at 2 min’s post-contrast. - AUC=0.69 for response prediction using entropy using pCR as reference |
| Michoux et al. [36] | 69 | 4 Cycles | GLCM (Semi-Automated segmentation) |
DCE-MRI-T1 (1 min after injection of contrast agent) |
Retro | pCR | - AUC=0.50 for 8 significant texture features - AUC=0.68 for 4 significant texure feature - AUC=0.74 for homogeneity feature as a best predictive model. |
| Ahmed et al. [38] | 100 | 4 Cycles | GLCM (Semi-Automated segmentation) |
DCE-MRI-T1 (1-5 min after injection of contrast agent) |
Retro | cR | - 8 texture parameters provided differences between response and partial response - High grade and TNBC present poorer prognosis with more heterogeneity. |
| Fox et al. [39] | 100 | 0 Cycles (baseline) |
Minkowski Functional (Semi-Automated segmentation) |
DCE-MRI-T1 (1 min after injection of contrast agent) |
Retro | cR | - Strong differences (P ≤ 0.01) were found between TNBC and other patient subgroups. - AUC=0.92 for TNBC classification |
| Parikh et al. [40] | 36 | 3 Cycles |
GLCM (Semi-Automated segmentation) |
DCE-MRI-T2 (1-3 min after injection of contrast agent) |
Retro | pCR | - Negative correlation between uniformity and entropy for all filters - AUC=0.84 was found 8 pCR and 8 cR. |
| Kim et al. [41] | 203 | Not mentioned | GLCM (Semi-Automated segmentation) |
DCE-MRI-T1 and subtraction T2 (90 s after injection of contrast agent) |
Retro | cR | - Mean cut-off=0.034 for T1-uniformity - Mean cut-off=5.057 for T1-entropy - Mean cut-off=0.019 for T2-uniformity - Mean cut-off=6.013 for T2-entropy |
9Gray Level Co-occurrence Matrix
Table 2: Literature on the texture analysis technique for assessing breast cancer response.
| Author | Patient | Timing | Method | Acquisition | Study1 | Reference | Finding |
|---|---|---|---|---|---|---|---|
| Cho et al. [43] | 48 | 1 Cycle |
Nonlinear Registration (Manual segmentation) |
DCE-MRI-T1 (7 min after injection of contrast agent) |
Prosp | pCR | - 6 patients achieved pCR and 42 showed npCR - AUC=0.71 for prediction of good response by PRM. |
| Boes et al. [44] | 100 | 4 Cycles | Registration Thin Plat Splines (Semi-Automated segmentation) |
DW-MRI-T1 & DW-MRI-T2 (4-5 min scan) |
Retro | pCR | - Description of PRM applied to breast tumor - Indication of significant changes in voxel-based changes in DW-MRI images using test-retest clinical scans. |
10 Complete Response
11 Triple-Negative Breast Cancer
12 Non-pathological response
■ Histogram analysis
Number of patient, timing, method finding and other information are presented in TABLE 1, where an overview of the research using histogram analysis as an index of breast cancer response to chemotherapy treatment is presented [28-30].
In this context, the skewness and kurtosis features extracted from tumors regions of patients with breast cancer receiving NAC were calculated in our institution. Histograms from MRI subtraction sequence of responsive (FIGURE 1) and non-responsive patients are presented in FIGURE 2.
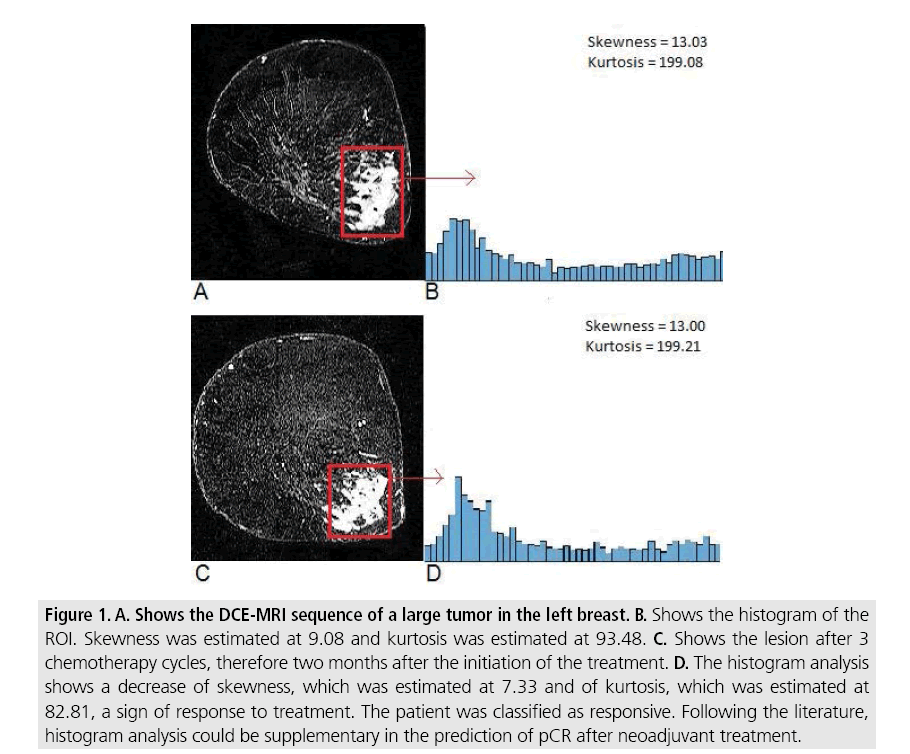
Figure 1: A. Shows the DCE-MRI sequence of a large tumor in the left breast. B. Shows the histogram of the ROI. Skewness was estimated at 9.08 and kurtosis was estimated at 93.48. C. Shows the lesion after 3 chemotherapy cycles, therefore two months after the initiation of the treatment. D. The histogram analysis shows a decrease of skewness, which was estimated at 7.33 and of kurtosis, which was estimated at 82.81, a sign of response to treatment. The patient was classified as responsive. Following the literature, histogram analysis could be supplementary in the prediction of pCR after neoadjuvant treatment.
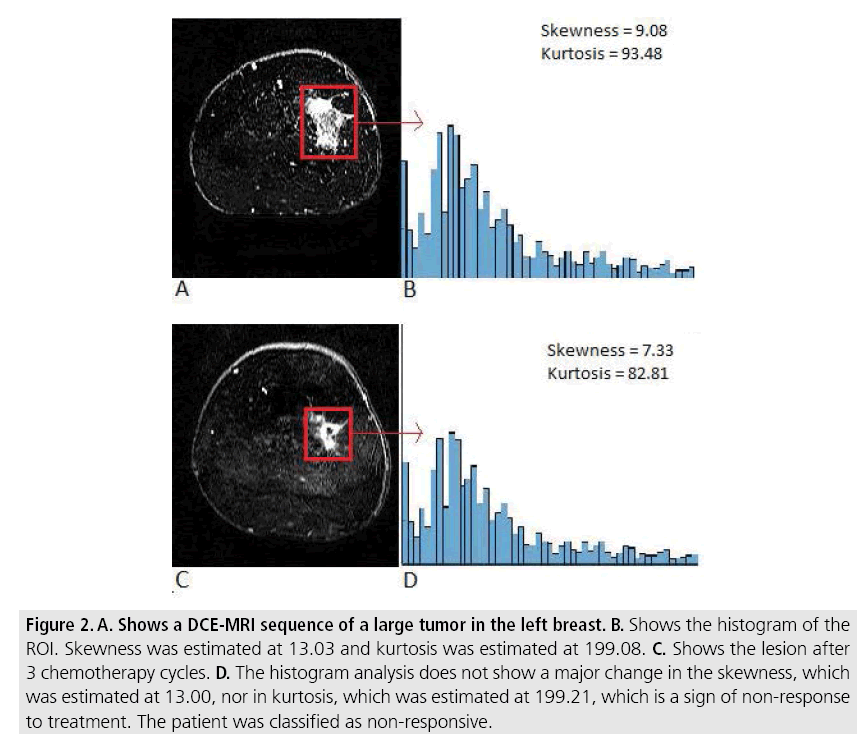
Figure 2: A. Shows a DCE-MRI sequence of a large tumor in the left breast. B. Shows the histogram of the ROI. Skewness was estimated at 13.03 and kurtosis was estimated at 199.08. C. Shows the lesion after 3 chemotherapy cycles. D. The histogram analysis does not show a major change in the skewness, which was estimated at 13.00, nor in kurtosis, which was estimated at 199.21, which is a sign of non-response to treatment. The patient was classified as non-responsive.
■ Texture analysis
In the context of texture analysis, we present in TABLE 2 an overview of researches using texture analysis as index to predict breast response to chemotherapy treatment.
■ Parametric response mapping (PRM)
TABLE 3 show the studies using PRM technique as tool for the prediction of breast tumor response to neoadjuvant treatment [44,45]. In this TABLE, Patient number, method and finding are summarized.
The PRM method is demonstrated using a data set for a patient with breast cancer receiving chemotherapy treatment in our institution. The obtained results are presented in FIGURE 3.
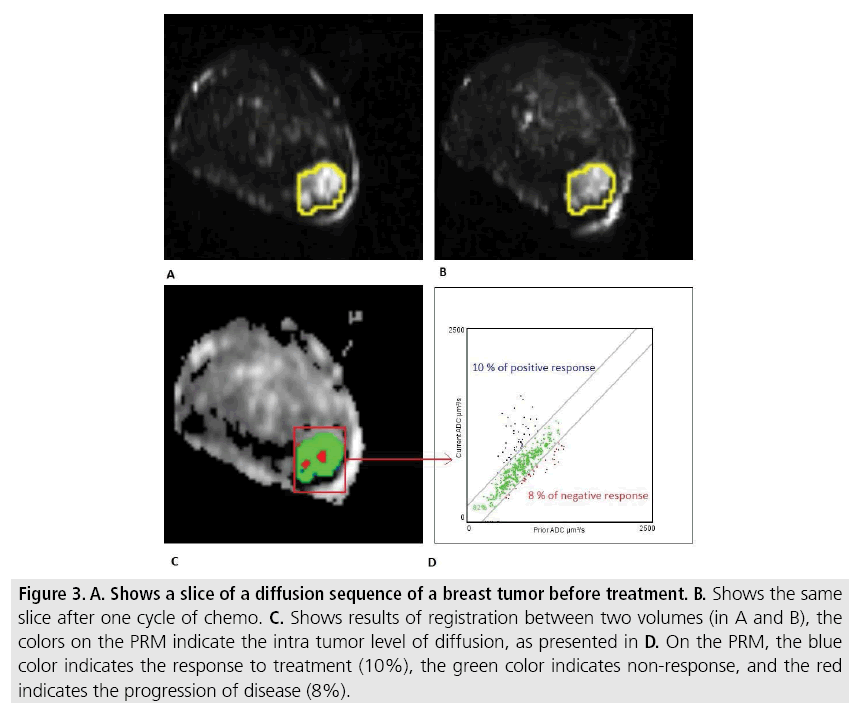
Figure 3: A. Shows a slice of a diffusion sequence of a breast tumor before treatment. B. Shows the same slice after one cycle of chemo. C. Shows results of registration between two volumes (in A and B), the colors on the PRM indicate the intra tumor level of diffusion, as presented in D. On the PRM, the blue color indicates the response to treatment (10%), the green color indicates non-response, and the red indicates the progression of disease (8%).
Discussion
In this paper, a review of literature related to the evaluation of the intra-tumor heterogeneity of breast tumors as a biomarker of treatment response was conducted. Three principal image analysis techniques were found: Histogram Analysis, Texture Analysis and Parametric Response Mapping (PRM). It was noted that most of the above-mentioned studies used 1 to 6 cycles of treatment [46-48].
Several histogram and texture based researches were focused on detection and identification of malignant and benign tumors. For example, Cho et al. [26] used histogram analysis to detect tumor malignancy level, Karahaliou et al. [46] investigated texture analysis to quantify the heterogeneity of breast lesions to discriminate malignant from benign ones. Chen et al. [47] and Nie et al. [49] extended the traditional GLCM method to investigate a volumetric texture analysis approach to characterize breast MRI lesions. Otherwise, there were fewer studies exploring the potential of intra-tumor heterogeneity as index of response to treatment.
In case of histogram analysis as index for predicting breast tumor response, it was concluded that a better approach is to gather relevant information in a set of descriptors. Nevertheless, among all the statics used for histogram analysis, no one has been found to be relevant to perform a complete index to assess breast tumor response to treatment. Histogram analysis as marker for breast tumor response was presented in the literature between 2002 and 2009 [28-31]. This method is relatively outdated and has only been validated with small number of patients (between 15 and 24).
For texture analysis, there are many important features to be calculated from GLCM matrix: entropy, uniformity, homogeneity, heterogeneity. For example, entropy represents the disorder and randomness of gray levels, and uniformity represents the gray-level distribution and determines how close values are uniform [28]. To achieve reliable results, some conditions should be considered such as, applying the technique of texture analysis to tumors with high-resolution and no discreet size. Texture analysis as marker for breast tumor response was presented in the literature between 2013 and 2016 [35-42] which is recent compared to histogram analysis. Additionally, the texture analysis technique was validated in the literature with a high number of patients (between 36 and 203).
Texture is defined as a repeating pattern, which has random variation. In fact, this method may be inappropriate for evaluating discreet tumors acquired in relatively low-resolution images, such as DW-MRI. Moreover, texture analysis can describe intra-tumor heterogeneity but cannot capture spatial modifications during treatment by the longitudinal comparison of baseline and post-treatment control examinations. An alternative strategy is to treat parameter maps as objects rather than textures. As highly heterogeneous parameter maps are more complex to analyze, the quantification of spatial heterogeneity can be obtained by determining the complexity of the parameter map objects [49].
Parametric Response Mapping (PRM) has been used primarily for brain cancer [50-52], where it has proven to be a relevant technique for predicting response to chemotherapy. Moffat et al. [52] used DCE-MRI acquisition for 20 patients with primary brain tumors. In this study, all MR images were spatially co-registered by using the pre-treatment T2-W sequence as the reference data set.
The variability in the numbers of studies for each imaging biomarker can be explained by the high number of parameters used in texture analysis, which confirms the results’ reliability. Therefore, histogram analysis does not give enough information for the relationship between the different pixels on the images. Only two studies were performed using PRM analysis for breast cancer response. One reason is that PRM relies heavily on breast image registration, which remains a challenging task due to low spatial resolution of DW-MRI, breast deformation during longitudinal scans and tumor size can change during chemotherapy treatment.
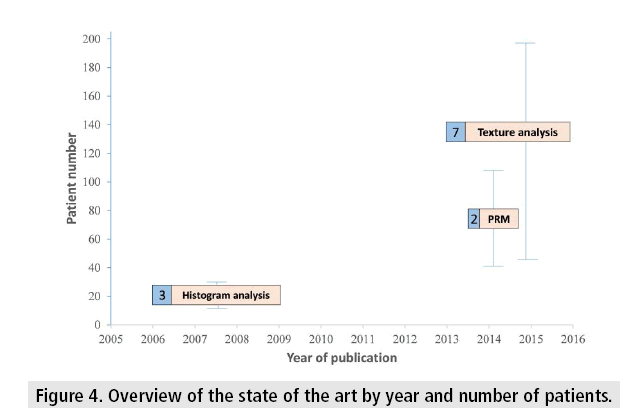
FIGURE 4 shows a comparison of the three methods presented in this paper, based on the number of patients, dates of publication, and the number of published papers.
The disadvantage that we can conclude based on this discussion is that the parameters used by each method differ from paper to another, depending on the type of MRI sub-modality and the statistical method used in each study. This can influence the choice of a standard or general method applied to all MRI breast data. So, based on the data used we can adapt technical methods such as Texture analysis to quantify a breast tumor heterogeneity that can provide a good prediction of response to treatment.
Conclusion and future work
Experimental evidence shows that MRI imaging provides an essential tool for assessing cancerous tumor evolution during NAC treatment. Several techniques have been developed to quantify and analyze breast tumor heterogeneity with DCE-MRI. In this paper, we presented different initial studies which deal with the imaging makers concept, applied to MRI sequences such DCE-MRI and DW-MRI, based on Histogram Analysis, Texture Analysis and PRM. These techniques have shown promising results for breast response assessment and tumor grading. This paper can be used as a guideline to help investigators to understand the above-mentioned concepts. Finally, PRM technique was introduced as an index of breast cancer response to NAC treatment, which has already been proven to be efficient in brain tumor assessment, and deserve more attention in future work for breast cancer.
Acknowledgement
Our thanks are addressed to Dr. Marc Lemort, the Head of Radiology department at Jule Bordet Institute in Brussels, for welcoming us and offering the Data set to evaluate our method. And we thank also Mr. Libert Gaetan the head of the Computer Science Unit in the Faculty of engineering in Mons.
Conflict of interest
All the authors declare that they have no conflict of interest to disclose.
Funding
This work was supported by the University of Mons in Belgium.
References
- Martelotto LG, Charlotte KY, Piscuoglio S et al. Breast cancer intra-tumor heterogeneity. Breast. Cancer. Res. 16, 210 (2014).
- Gerlinger M, Rowan AJ, Horswell S et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883-892 (2012).
- Vin N, Krasnitz A, Rodgers L et al. Inferring tumor progression from genomic heterogeneity. Genome. Res. 20, 68-80 (2010).
- Navin N, Kendall J, Troge J et al. Tumour evolution inferred by single-cell sequencing. Nature. 472, 90-94 (2011).
- Shah SP, Morin RD, Khattra J et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 461, 809-813 (2009).
- Bedard PL, Hansen AR, Ratain MJ et al. Tumour heterogeneity in the clinic. Nature. 501, 355-364 (2013).
- Geyer FC, Weigelt B, Natrajan R, et al. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J. Pathol. 220, 562-573 (2010).
- Eisenhauer E, Therasse P, Bogaerts J et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer 45, 228-247 (2009).
- Fisher B, Bryant J, Wolmark N et al. Effect of preoperative chemotherapy on the outcome of women with operable breast cancer. J. Clin. Oncol. 16, 2672-2685 (1998).
- Therasse P, Mauriac L, Welnicka-Jaskiewicz et al. Final results of a randomized phase III trial comparing cyclophosphamide, epirubicin and fluorouracil with a dose-intensified epirubicin and cyclophosphamide+filgrastim as neoadjuvant treatment in locally advanced breast cancer: An EORTC-NCIC-SAKK multicenter study. J. Clin. Oncol. 21, 843-850 (2003).
- Jemal A, Bray F, Center MM et al. Global cancer statistics. CA. Cancer. J. Clin. 61, 69-90 (2011).
- Polyak K. Heterogeneity in breast cancer. J. Clin. Invest. 121, 3786-3788 (2011).
- Agrawal G, Su MY, Nalcioglu O et al. Significance of breast lesion descriptors in the ACR BI‐RADS MRI lexicon. Cancer. 115, 1363-1380 (2009).
- Kaufmann M, Minckwitz V, Mamounas G et al. Recommendations from an international consensus conference on the current status and future of neoadjuvant systemic therapy in primary breast cancer. Ann. Surg. Oncol. 19, 1508-1516 (2012).
- Wolmark N, Wang J, Mamounas E et al. Preoperative chemotherapy in patients with operable breast cancer: Nine year results from National Surgical Adjuvant Breast and Bowel Project B-18. J. Natl. Cancer. Inst. Monogr. 30, 96-102 (2001).
- Esserman L, Hylton N, Yassa L et al. Utility of magnetic resonance imaging in the management of breast cancer: Evidence for improved preoperative staging. J. Clin. Oncol. 17, 110-110 (1999).
- Weatherall PT, Evans GF, Metzger GJ et al. MRI vs. histologic measurement of breast cancer following chemotherapy: Comparison with x‐ray mammography and palpation. J. Magn. Reson. Imaging 13, 868-875 (2001).
- Schnall MD, Blume J, Bluemke DA et al. MRI detection of distinct incidental cancer in women with primary breast cancer studied in IBMC 6883. J. Surg. Oncol. 92, 32-38 (2005).
- Van Goethem M, Schelfout K, Dijckmans L et al. MR mammography in the pre-operative staging of breast cancer in patients with dense breast tissue: comparison with mammography and ultrasound. Eur. Radiol. 14, 809-816 (2004).
- Berg WA, Gutierrez L, NessAiver MS et al. Diagnostic accuracy of mammography, clinical examination, US and MR imaging in preoperative assessment of breast cancer 1. Radiology. 233, 830-849 (2004).
- Liney GP, Gibbs P, Hayes C et al. Dynamic contrast‐enhanced MRI in the differentiation of breast tumors: User‐defined versus semi‐automated region‐of‐interest analysis. J. Magn. Reson. Imaging. 10, 945-949 (1999).
- Kang H, Lee HY, Lee KS et al. Imaging-based tumor treatment response evaluation: review of conventional, new and emerging concepts. Korean. J. Radiol. 13, 371-390 (2012).
- Campbell LL, Polyak K. Breast tumor heterogeneity: Cancer stem cells or clonal evolution? Cell. Cycle. 6, 2332-2338 (2007).
- Just N. Improving tumour heterogeneity MRI assessment with histograms. Br. J. Cancer. 111, 2205-2213 (2014).
- Issa B, Buckley DL, Turnbull LW. Heterogeneity analysis of Gd-DTPA uptake: improvement in breast lesion differentiation. J. Comput. Assist. Tomogr. 23, 615-621 (1999).
- Cho GY, Moy L, Kim SG et al. Evaluation of breast cancer using intravoxel incoherent motion (IVIM) histogram analysis: comparison with malignant status, histological subtype and molecular prognostic factors. Eur. Radiol. 26, 2547-2558 (2016).
- Carter JS, Koopmeiners JS, Kuehn‐Hajder JE et al. Quantitative multiparametric MRI of ovarian cancer. J. Magn. Reson. Imaging. 38, 1501-1509 (2013).
- Johansen R, Jensen LR, Rydland J, et al. Predicting survival and early clinical response to primary chemotherapy for patients with locally advanced breast cancer using DCE‐MRI. J. Magn. Reson. Imaging. 29, 1300-1307 (2009).
- Padhani AR, Hayes C, Assersohn L, et al. Leach and Janet E. Husband. Prediction of clinicopathologic response of breast cancer to primary chemotherapy at contrast-enhanced MR imaging: Initial clinical results 1. Radiology. 239, 361-374 (2006).
- Li L, Wang K, Sun X et al. Parameters of dynamic contrast-enhanced MRI as imaging markers for angiogenesis and proliferation in human breast cancer. Med. J. Exp. Clin. Res. 21, 376 (2015).
- Hayes C, Padhani AR, Leach MO. Assessing changes in tumour vascular function using dynamic contrast‐enhanced magnetic resonance imaging. NMR. Biomed. 15, 154-163 (2002).
- Whelan PF, Molloy D. Machine vision algorithms in Java: Techniques and implementation. Springer. New. York. Science. & Business. Media. (2001).
- Haralick RM, Shanmugam K. Textural features for image classification. IEEE Trans. Syst. Man. Cybern. Syst. 3, 610-621 (1997).
- Gunjan M, Arpitam C, Bipan T. Study on the potential of combined GLCM features towards medicinal plant classification. In control, instrumentation, energy & communication (CIEC) 2nd International Conference on IEEE 98-102 (2016).
- Golden DI, Lipson JA, Telli ML et al. Dynamic contrast-enhanced MRI-based biomarkers of therapeutic response in triple-negative breast cancer. J. Am. Med. Inform. Assoc. 20, 1059-1066 (2013).
- Teruel JR, Heldahl MG, Goa PE et al. Dynamic contrast‐enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR. Biomed. 27, 887-896 (2014).
- Michoux N, Broeck V, Lacoste S et al. Texture analysis on MR images helps predicting non-response to NAC in breast cancer. BMC. Cancer. 15, 574 (2015).
- Tang X. Texture information in run-length matrices. IEEE Trans. Image. Process. 7, 1602-1609 (1998).
- Ahmed A, Gibbs P, Pickles M et al. Texture analysis in assessment and prediction of chemotherapy response in breast cancer. J. Magn. Reson. Imaging. 38, 89-101 (2013).
- Fox MJ, Gibbs P, Pickles MD. Minkowski functionals: An MRI texture analysis tool for determination of the aggressiveness of breast cancer. J. Magn. Reson. Imaging. 43, 903-910 (2015).
- Parikh J, Selmi M, Charles-Edwards G et al. Changes in primary breast cancer heterogeneity may augment mid-treatment MR imaging assessment of response to neoadjuvant chemotherapy. Radiology. 272, 100-112 (2014).
- Kim JH, Ko ES, Lim Y et al. Breast cancer heterogeneity: MR imaging texture analysis and survival outcomes. Radiology. 282, 665-675 (2017).
- Galbán CJ, Chenevert TL, Meyer CR et al. The parametric response map is an imaging biomarker for early cancer treatment outcome. Nat. Med. 15, 572-576 (2009).
- Cho N, Im SA, Park IA et al. Breast cancer: Early prediction of response to neoadjuvant chemotherapy using parametric response maps for MR imaging. Radiology. 272, 385-396 (2014).
- Boes JL, Hoff BA, Hylton N et al. Image registration for quantitative parametric response mapping of cancer treatment response. Transl. Oncol. 7, 101-110 (2014).
- Karahaliou A, Vassiou K, Arikidis NS et al. Assessing heterogeneity of lesion enhancement kinetics in dynamic contrast-enhanced MRI for breast cancer diagnosis. Br. J. Radiol. 83, 296-309 (2010).
- Chen W, Giger ML, Li H et al. Volumetric texture analysis of breast lesions on contrast‐enhanced magnetic resonance images. Magn. Reson. Med. 58, 562-571 (2007).
- Nie K, Chen J, Hon H et al. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad. Radiol. 15, 1513-1525 (2008).
- Rose CJ, Mills SJ, O'Connor JP et al. Quantifying spatial heterogeneity in dynamic contrast‐enhanced MRI parameter maps. Magn. Reson. Med. 62, 488-499 (2009).
- Galbán CJ, Chenevert TL, Meyer CR et al. Prospective analysis of parametric response map-derived MRI biomarkers: identification of early and distinct glioma response patterns not predicted by standard radiographic assessment. Clin. Cancer. Res. 17, 4751-4760 (2011).
- Hamstra DA, Galbán CJ, Meyer CR et al. Functional diffusion map as an early imaging biomarker for high-grade glioma: Correlation with conventional radiologic response and overall survival. J. Clin. Oncol. 26, 3387-3394 (2008).
- Moffat BA, Chenevert TL, Lawrence TS et al. Ross functional diffusion map: a noninvasive MRI biomarker for early stratification of clinical brain tumor response. Proc. Natl. Acad. Sci. USA 102, 5524-5529 (2005).