Research Article - Journal of Medicinal and Organic Chemistry (2022) Volume 5, Issue 5
Antimicrobial peptides and their interaction Cell Membrane and Microbial Surface Interactions
Ahmed Tanvir*
Institute of Molecular Biosciences, Biophysics Division, University of Graz, NAWI Graz, Austria; BioTechMed-Graz, Austria
Institute of Molecular Biosciences, Biophysics Division, University of Graz, NAWI Graz, Austria; BioTechMed-Graz, Austria
E-mail: Tanvirahmed@edu.org
Received: 04-Oct-2022, Manuscript No. JMOC-22-74668; Editor assigned: 06-Oct-2022, PreQC No. JMOC-22- 74668 (PQ); Reviewed: 18-Oct-2022, QC No. JMOC-22-74668; Revised: 24-Oct-2022, Manuscript No. JMOC- 22-74668 (R); Published: 31-Oct-2022 DOI: 10.37532/jmoc.2022.5(5).86-92
Abstract
Antimicrobial peptides (AMPs) selectively kill bacteria by disrupting their cell membranes, and are promising compounds to fight drug-resistant microbes. Biophysical studies on model membranes have characterized AMP/membrane interactions and the mechanism of bilayer perturbation, showing that accumulation of cationic peptide molecules in the external leaflet leads to the formation of pores. However, similar quantitative studies on real cells are extremely limited. Here, we investigated the interaction of the dansylated PMAP23 peptide with a Gram-positive bacterium, showing that 107 bound peptide molecules per cell are needed to kill it. This result is consistent with our previous finding for Gram-negative strains, where a similar high threshold for killing was determined, demonstrating the general relevance of the carpet model for real bacteria. However, in the case of the Gram-positive strain, this number of molecules even exceeds the total surface available on the bacterial membrane. The high affinity of DNS-PMAP23 for the anionic teichoic acids of the Gram-positive cell wall, but not for the lipopolysaccharides of Gram-negative bacteria, provides a rationale for this finding. To better define the role of anionic lipids in peptide/cell association, we studied DNS-PMAP23 interaction with E. coli mutant strains lacking phosphatidylglycerol and/or cardiolipin. Surprisingly, these strains showed a peptide affinity similar to that of the wild type. This finding was rationalized by observing that these bacteria have an increased content of other anionic lipids, thus maintaining the total membrane charge essentially constant. Finally, studies of DNS-PMAP23 association to dead bacteria showed an affinity an order of magnitude higher compared to that of live cells, suggesting strong peptide binding to intracellular components that become accessible after membrane perturbation. This effect could play a role in population resistance to AMP action, since dead bacteria could protect the surviving cells by sequestering significant amounts of peptide molecules. Overall, our data indicate that quantitative studies of peptide association to bacteria can lead to a better understanding of the mechanism of action of AMPs.
Introduction
The emergence and diffusion of drug resistant bacteria is one of the greatest health challenges of our time. We are currently at risk of losing the transformative medical progress made possible by antibiotic drugs, such as the easy treatment of deadly infectious diseases, surgery, transplants or anticancer chemotherapy. The latest report on antibiotic resistance by the US Centers for Disease Control and Prevention warns us that we are already living in a post-antibiotic era: in the US alone, one person is killed by drug resistant microbes every 15 min [1]. Global deaths by “superbugs”, as these bacteria are called, are currently estimated at >700,000 per year, but this figure could increase to 10 million by 2050 if no action is taken. New drugs against resistant bacteria are severely needed.
Antimicrobial peptides are an important component of our innate defense system. These molecules have multiple functions, including immunomodulatory roles, but they are mainly bactericidal. Thousands of different natural AMPs are currently known: their sequences do not show any conserved motif, but they all share some properties. They are usually short, cationic and amphipathic. Most AMPs act by binding to bacterial membranes and making them permeable, through the formation of pores [2]. Since this mechanism of action does not involve any specific protein target, development of bacterial resistance is difficult and AMPs have a wide spectrum of activity and are effective also against resistant strains.
AMPs have the additional desirable property of being moderately selective, i.e. they are active against bacteria at concentrations where their toxicity to the host cells is negligible. This property is attributed mostly to the fact that microbial membranes display lipids with negatively charged phospholipid headgroups on their surface, while in the membranes of eukaryotic cells anionic lipids are segregated to the inner leaflet. Consistently, AMPs interact more strongly with anionic than with neutral artificial membranes, due to their cationic nature [3].
Studies on model membranes led to the proposal of several mechanisms of pore formation by AMPs, but the most widely applicable model is based on accumulation of peptides on the outer leaflet of the bilayer, perturbation of its surface tension and formation of pores or defects. In artificial vesicles, the bound peptide to lipid ratios necessary for perturbation of membrane permeability are typically of the order of 1/10, which corresponds to complete coverage of the bilayer. For this reason, this mechanism has been termed the “carpet” model.
One crucial issue is whether the behavior, now observed only for a very limited number of AMPs and bacterial strains, is relevant also for other systems. In particular, to the best of our knowledge, only one experiment on Grampositive bacteria has been reported, on the association of cecropin A to B. megaterium. However, this bacterium is rather atypical due to its unusual size [4], as it can reach up to 4 μm of length. Considering the great differences in the cellular envelopes between Gram-negative and Gram-positive bacteria, further studies focusing on Gram-positives are warranted.
The very high accumulation of peptides needed for killing begs the question of where all these molecules are located in a bacterial cell. A related ongoing debate also regards the role of the saccharidic portion of lipopolysaccharide (LPS) and of cell wall components in the mechanism of action of AMPs. Do these components facilitate the activity of AMPs, by guiding their access to the plasma membrane [5], or do they rather inhibit their action by sequestering peptide molecules and reducing their effective concentration? Reviews on this topic by Prof. Karl Lohner outlined this issue and urged for further affinity studies of AMPs towards LPS and cell wall components, inspiring us to pursue our investigations further.
Finally, three microscopy studies, published in the last year, reported an accumulation of AMPs inside the bacterial cells, after membrane permeabilization. This phenomenon might be just the consequence of the accessibility of internal molecules and components as additional binding partners, or it could contribute to bacterial killing. In any case, peptide sequestration by dead cells might help other bacteria to survive to AMP action, by reducing the available peptide concentration [6].
With the goal of addressing these questions, we have investigated the interaction of DNSPMAP23 with a Gram-positive bacterium, with lipoteichoic acids, with bacterial cells in which specific mutations caused variations in the LPS structure or in the lipid composition of the cytoplasmic membrane, and with live and dead bacterial cells.
Materials and Methods
Materials
DNS–PMAP23 was purchased from AnyGen Co. Bacterial culture media were purchased from Oxoid, 96–well plates were from Falcon. Lipoteichoic acid (LTA) was purchased from Sigma Aldrich.
Microorganisms
The following strains were used for the antimicrobial assays: the Gram-negative bacteria Escherichia coli ATCC 25922, D21 and E. coli D21f2, a D21 mutant in genes rfa-1 and rfa-31 lacking of the O-antigen and oligosaccharide core, E. coli MG1655, and its mutant strains BKT12 and BKT29 (lacking the pgsA gene, in addition to the clsABC and ymdB genes [7]; pgsA encodes a phosphatidylglycerol phosphate synthase, which catalyzes PG synthesis, from phosphatidic acid; the Gram-positive bacterium Staphylococcus epidermidis ATCC 12228. Strains BKT12 and BKT29 were a kind gift by Prof. J. C. Weisshaar.
Antimicrobial Assays
Bactericidal Activity
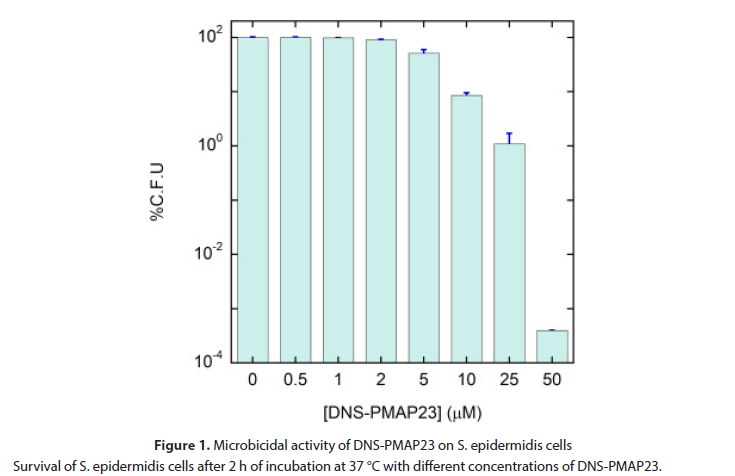
With the goal of determining the bactericidal activity of DNS-PMAP23, in the presence of a well-defined number of S. epidermidis cells, the following assay was developed, similarly to our previous studies on E. coli. S. epidermidis was grown in Luria Bertani (LB) medium at 37 °C under mild agitation (200 rpm), to a mid-logarithmic phase. After centrifugation at 1400 ×g for 10 min and washing for three times [8], the bacterial culture was resuspended in buffer A, a minimal culture medium where bacteria are vital, but do not multiply, thus maintaining a constant number of live cells in the timeframe of the experiments (Figure 1).
Figure 1: Microbicidal activity of DNS-PMAP23 on S. epidermidis cells Survival of S. epidermidis cells after 2 h of incubation at 37 °C with different concentrations of DNS-PMAP23.
Determination of Minimum Inhibitory Concentration (MIC)
In addition to the bactericidal activity, MIC was determined, too, for comparison, by adapting the microbroth dilution method, using sterile 96-wells plates. Aliquots (50 μL) of bacteria in mid-logarithmic phase at a concentration of 2×106 CFU/mL in culture medium were added to 50 μL of MH broth containing the peptide in serial 2-fold dilutions in methanol [9]. The MIC was expressed as the minimum concentration of peptide at which 100% inhibition of microbial growth is visually observed after 16–18 h of incubation at 37 °C.
Preparation of Dead Bacteria
To measure peptide association to dead bacterial cells, bacteria grown to the midlogarithmic phase were centrifuged at 1400 ×g for 10 min and washed eight times; afterwards, they were resuspended in buffer A at a final cell density of 6 × 109 CFU/mL. Two mL were then sonicated with Sonic Vibra Cell to kill bacteria [10].
Zeta-Potential Measurements
Peptide/cell association was characterized also by measurements of zeta potential. Midlogarithmic growing cells were diluted to 1 × 107 CFU/mL in Hepes buffer and exposed to different peptide concentrations. After 5 min incubation at room temperature (RT), they were measured on a Zetasizer according to previously published methods [11].
Lipid Analysis
The membrane lipid composition of mutant E. coli strains BKT12 and BKT29, and of the parent strain MG1655, was characterized by lipid analysis. Bacteria were grown overnight at 37 °C in MH broth and then inoculated at 0.05 optical density (OD) to a fresh MH broth [12]. The growth was stopped after 4 h, corresponding to the mid-logarithmic growth phase, and cells were collected by centrifugation. Approximately 10 mL of a 1 OD cell suspension were washed twice with phosphate buffered saline, pH = 7 and kept at-80 °C, until further preparation steps.
Results and Discussion
Extending our previous studies on E. coli, we investigated the interaction of DNSPMAP23 with the Gram-positive bacterium S. epidermidis.
MIC of DNS-PMAP23 against the Gramnegative E. coli (ATCC 25922) and the Grampositive S. epidermidis were comparable, in agreement with previous reports on the activity of the unlabeled peptide against the two bacterial classes. However, the MIC assay was not suitable for our goal of determining the number of cell-bound peptide molecules needed for bacterial killing [13]. By definition, the MIC assay requires a medium in which bacteria grow, while we needed a welldefined and stable cell density. In addition, the culture broth used in the MIC assay can sequester a significant amount of peptide and interfere with spectroscopic measurements.
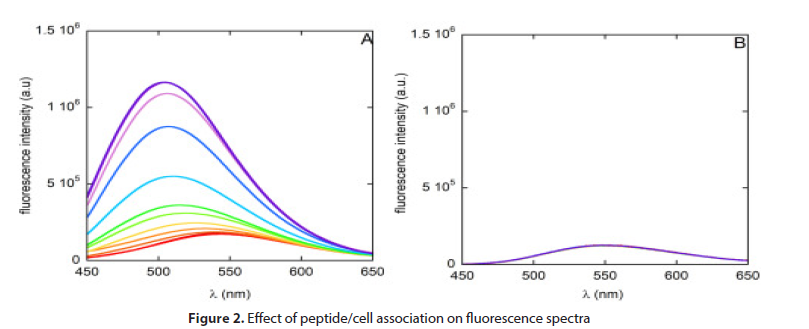
.We performed experiments of peptide binding to bacterial membranes by fluorescence spectroscopy. PMAP-23 is intrinsically fluorescent, but the introduction of an extrinsic dansyl fluorophore at the N-terminus of DNS-PMAP23 was necessary to obtain a signal not overlapping to the intrinsic fluorescence of bacterial proteins. Although a slight perturbation of the peptide behavior by the fluorophore is possible, by performing binding and killing experiments under the same conditions of the MBC assays, we ensured that the probe affected both datasets in the same way, so that it could be factored out.
Titration of DNS-PMAP23 with increasing densities of S. epidermidis cells caused an increase in the fluorescence intensity and a blue shift of the maximum of the emission spectrum, as previously observed with E. coli. These are distinctive features of dansyl insertion in an apolar, relatively rigid environment and thus indicate association to bacterial membranes [14]. By contrast, performing a similar titration with the dansyl analogue dansylamide did not cause significant variations in the fluorescence signal, ruling out possible artifacts due to sample turbidity, caused by the bacterial cells in suspension (Figure 2).
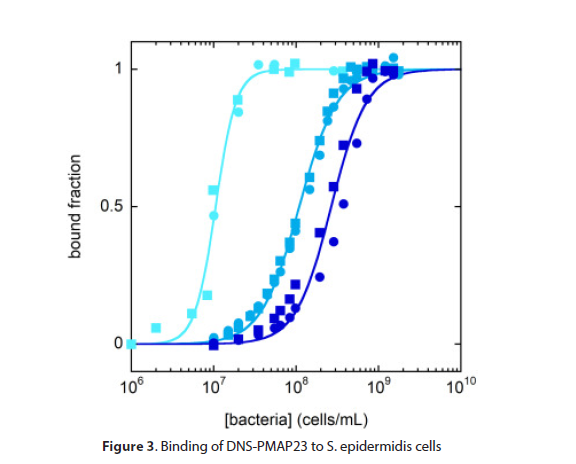
We calculated the fraction of peptide bound to bacterial cells from the variation of fluorescence intensity at a fixed wavelength (515 nm) (Figure 3). Apparent dissociation constants determined from these curves are reported in (Table 1). Their order of magnitude is comparable to the values previously determined for E. coli. The dependence of the curves on peptide concentration is significantly more marked than in the case of E. coli cells. This finding is compatible with an association process where electrostatic interactions play a significant role [15], rather than with an ideal water/membrane partition equilibrium, and is consistent with the fact that the content of anionic lipids in Gram-positive bacterial membranes is significantly higher than in Gram-negatives.
Table 1.Membrane coverage by DNS-PMAP-23 under the conditions of the bactericidal assayBacterium |
[DNS-PMAP23] (μM) | Fraction of surviving bacteriaa | C50(cells/mL)b | Bound fractionc | Bound molecules/celld | Ac/Ame |
|---|---|---|---|---|---|---|
| S. epidermidisATCC 12228 (Gram-positive) | 2 | 0.91±0.02 | (1.05±0.004)×107 | 1 | (2.7±0.3)×106 | 1–8 |
| 10 | 0.09±0.01 | (1.09±0.02)×108 | 0.98±0.03 | (13±1)×106 | 5–40 | |
| 50 | (4±1)×10−6 | (2.7±0.1)×108 | 0.8±0.1 | (50±10)×106 | 20–150 | |
| E. coliATCC 25922 (Gram-negative)f | 1 | 0.94±0.03 | (9.0±0.9)×107 | 0.89±0.08 | (1.2±0.2)×106 | 0.2 |
| 10 | (5±7)×10−4 | (1.8±0.2)×108 | 0.829±0.003 | (11±1)×106 | 2 |
Conclusion
In this work, we have extended our previous studies on the quantitative determination of AMP binding to bacterial cells. Together with the determination of bacterial killing under the same conditions, these experiments provided several novel insights in the mechanism of action of AMPs.
We found that, for both Gram-negative and Gram-positive bacteria, ~107 AMP molecules must accumulate on a single cell to kill it. This number is supported from the few studies available in the literature, in addition to ours, using different techniques, AMPs and bacteria. In particular, here we extended its validity to Gram-positive microbes. Therefore, this finding seems to be of general relevance.
The huge amount of peptide molecules needed to cause cell death provides strong support to the carpet model for the mechanism of pore formation in bacterial membranes. Since this model was originally devised based on physico-chemical studies on artificial bilayers, its generalization to real cells underlines the importance of quantitative investigation of AMP-membrane interactions, and indicates that liposomes are a useful model to investigate the mechanism of action of AMPs, despite their simplicity. In particular, it had been argued that the bilayer coverage needed for pore formation in artificial membranes was too high to be reached at the concentrations needed for bacterial killing. On the contrary, our data demonstrate that the high affinity of AMPs for bacteria explains why this is actually happening under the conditions of in vitro assays.
In the case of the Gram-positive bacteria studied here, the number of ~107 molecules even exceeds the total available surface area. Determination of the binding affinity of DNS-PMAP23 for LTA indicated that peptide association to this cellular component, and sequestration of a significant fraction of the peptide molecules, could explain this finding. By contrast, in the case of Gram-negative bacteria, the core region of LPS appears to have a marginal role in peptide binding, possibly providing a rationale for the lower membrane coverage needed in the case of those cells. On the other hand, we observed that truncation of the core portion increases the susceptibility of bacteria to AMP action, possibly due to the loss of the stabilizing effect of the saccharides on the structure of the outer membrane. Our findings contribute to the ongoing debate on the role of LPS in the action of AMPs, indicating that stabilization of the outer membrane, rather than peptide sequestration, can explain its protective action.
The anionic charge of bacterial membranes is generally considered one of the principal determinants of the selectivity of AMPs for pathogen versus host cells. However, this idea is based mainly on studies on artificial membranes. We tried to verify it in real bacteria by exploiting bacterial strains where the synthesis of PG and/or CL was blocked by specific mutations. Surprisingly, elimination of PG or of PG and CL did not cause a reduction in AMP/cell association. Lipidomic studies showed that this unexpected result was due to the compensation of the lacking lipids by other anionic membrane components. This finding underlines the difficulty involved in the development of bacterial resistance to AMPs by alteration of the membrane composition and charge.
Finally, our data showed that, once the cell membranes lose their integrity, AMP can access intracellular cell components and peptide/cell association increases by an order of magnitude. This finding provides a quantitative justification for the recent observation of AMP accumulation in dead bacterial cells and could be relevant to better describe bacterial population susceptibility to AMPs. Peptide sequestration by dead cells reduces the available peptide concentration and can thus protect the remaining bacteria from the action of AMPs. Our result, by determining the affinity for both live and dead bacterial cells, can provide a basis for quantitative modeling of this effect.
Of course, several questions remain open regarding the interaction of AMPs with bacterial cells. The application of quantitative physico-chemical and biophysical approaches to studies of peptide/cell association is a powerful tool for the characterization of AMPs that will hopefully lead to a step forward in our understanding of these molecules and in their development for therapeutic purposes.
References
- Van der Does AM,Hiemstra PS,Mookherjee N. Antimicrobial host defence peptides: immunomodulatory functions and translational prospects. Adv Exp Med Biol.1117,149-171(2019).
- S.Bobone,L.Stella. Selectivity of antimicrobial peptides: a complex interplay of multiple equilibria. Adv Exp Med Biol.1117, 175-214 (2019).
- Matsuzaki K. Membrane permeabilization mechanisms. Adv Exp Med Biol.1117,9-16 (2019).
- Melo MN, Ferre Castanho R. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat Rev Microbiol.7,245-247 (2019).
- Gazit E, Miller IR, Biggin PC et al. Structure and orientation of the mammalian antibacterial peptide cecropin P1 within phospholipid membranes. J Mol Biol.258,860-870 (1996).
- Roversi D, Luca V, Aureli S et al. How many antimicrobial peptide molecules kill a bacterium? The case of PMAP-23. ACS Chem Biol.9,2003-2007 (2014).
- Savini F, Luca V, BocediR et al. Cell-density dependence of host-defense peptide activity and selectivity in the presence of host cells. ACS Chem Biol.12,52-56 (2012).
- Savini F,Bobone S, Roversi D et al. From liposomes to cells: filling the gap between physicochemical and microbiological studies of the activity and selectivity of host-defense peptides. Pept Sci.110, 345-349(2018).
- Snoussi M, Talledo JP,Del Rosario NA et al. Heterogeneous absorption of antimicrobial peptide LL37 in Escherichia coli cells enhances population survivability. E Life.7, 45-48(2018).
- Starr CG, He J, Wimley WC et al. Host cell interactions are a significant barrier to the clinical utility of peptide antibiotics. ACS Chem Biol.11, 3391-3399 (2016).
- Zhu Y, Mohapatra S, Weisshaar JC et al. Rigidification of the Escherichia coli cytoplasm by the human antimicrobial peptide LL-37 revealed by super resolution fluorescence microscopy. Proc Natl Acad Sci.116,1017-1026 (2019).
- Wu F, Tan C. Dead bacterial absorption of antimicrobial peptides underlies collective tolerance. J R Soc Interface.16, 3457-3459 (2019).
- Bocchinfuso G, Palleschi A, Orioni B et al. Different mechanisms of action of antimicrobial peptides: insights from fluorescence spectroscopy experiments and molecular dynamics simulations. J Pept Sci. 15,550-558 (2009).
- Orioni B, Bocchinfuso G, Kim JY et al. Membrane perturbation by the antimicrobial peptide PMAP-23: a fluorescence and molecular dynamics study. Biochim Biophys Acta – Biome.1788,1523-1533 (2009).
- Steiner H, Andreu D, Merrifield RB et al. Binding and action of cecropin and cecropin analogues: antibacterial peptides from insects. Biochim Biophys Acta – Biome.939,260-266 (1998).
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref