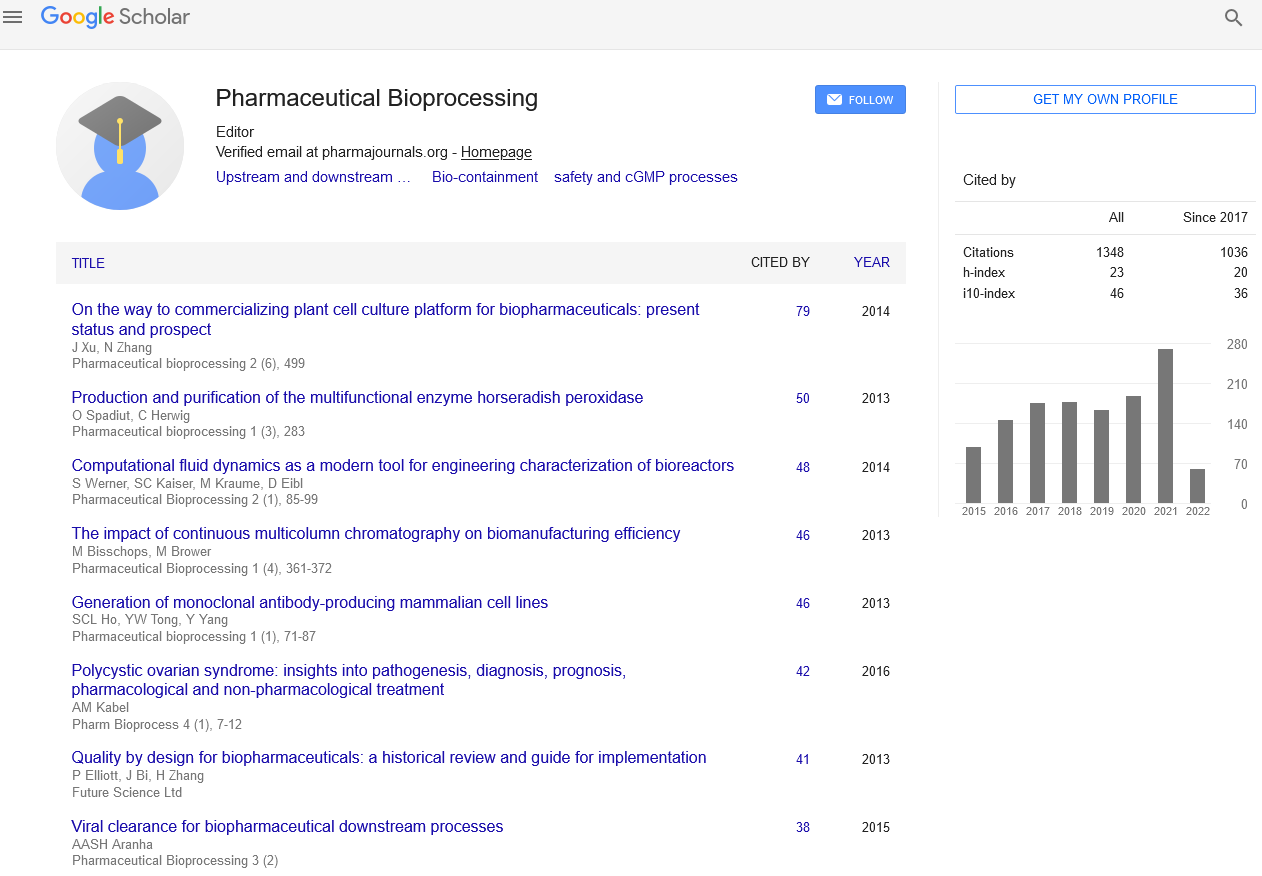
Editorial - Pharmaceutical Bioprocessing (2016) Volume 4, Issue 5
Considerations on the Use of Enzymes in the Downstream Processing of Biopharmaceuticals
- Corresponding Author:
- Duarte Miguel F Prazeres
iBB- Institute for Bioengineering and Biosciences, Department of Bioengineering, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, 1049-001 Lisboa, Portugal
E-mail: miguelprazeres@tecnico.ulisboa.pt
Abstract
Keywords
hypertension, antihypertensive drugs, adherence, efficacy
Enzyme-assisted process steps
Enzyme-assisted process steps can be found occasionally in the downstream processing of highly valuable biopharmaceuticals (Table 1). The implementation of such a step will typically involve the addition of a certain amount of a purified enzyme preparation to a productcontaining stream at a certain stage of the process. The step is designed to take advantage of the catalytic activity of an enzyme in vitro to (i) facilitate cell disruption or cell dissociation, (ii) promote the degradation of cell derivedimpurities or (iii) perform very specific alterations in the structure of key biomolecules.
| Category | Application | Enzyme | Ref. |
|---|---|---|---|
| Cell lysis and dissociation | Lysis and disruption of microbial cells for intracellular product release | Lysozyme, glucanases, mannanases, chitinases | [1-4] |
| Detachment of adherent cells of from culture vessels for passage | Trypsin | [5-8] | |
| Impurity clearance | Clearance of nucleic acids from process streams arising from mammalian cell culture | DNase | [12-18] |
| Digestion of plasmid DNA vectors following transient transfection of cells in viral particle production | DNase | [19,20] | |
| Clearance of host cell (E. coli) RNA in plasmid DNA purification | RNase | [21-23] | |
| Digestion of linear DNA fragments in plasmid DNA purification | Exonuclease | [25] | |
| Targeted structural modifications | Removal of affinity or solubility tags from protein fusions via hydrolysis ofpeptidic bonds | Trypsin, TEV, Factor Xa, enteropeptidase, R3C, thrombin,DAPase,carboxypeptidase A and B | [26-27] |
| Generation of insulin via cleavage of recombinant proinsulin | Trypsin, carboxypeptidase B |
[31-33] | |
| Generation of antibody fragments by proteolytic digestion of whole antibodies | Papain, pepsin | [34-38] | |
| Glycosylation of recombinant proteins produced in prokaryotic hosts | Endo-glycosidases | [39-43] | |
| Linearization of miniplasmid impurities in minicircle vector purification | Restriction endonuclease | [44] | |
| Modification of topology of miniplasmid impurities in minicircle vector purification | Nicking endonuclease | [45] |
Table 1: Examples of enzyme-assisted process steps in the downstream processing of biopharmaceuticals. Abbreviations: TEV (tobacco etch virus protease), R3C (rhinovirus 3C protease).
Cell lysis and dissociation
The enzymatic lysis and disruption of microbial cells as an early step of product isolation is well established and dates back to the early years of modern Biotechnology [1]. Enzymes like glucanases, mannanases, chitinases, glycosidases and endopeptidases have been studied at lab scale, but lysozyme is by far the most important lytic enzyme for process scale disruption of cells [1,2]. The ability of this enzyme to hydrolyze β-1,4-glycosidic bond make it an excellent choice to disrupt bacterial cells walls containing peptidoglycan, especially when used in combination with chelating agents like EDTA [3] or in conjunction with disruption methods like wet milling and high pressure homogenization [4].
Another well-known application of enzymes in the early downstream processing is found in the harvesting and sub-culturing of mammalian cells. These operations rely heavily on trypsin, a proteolytic enzyme that breaks down proteins to detach adherent cells from culture vessels for passaging [5]. Trypsinization is common in the manufacture of vaccines [6], advanced therapy medicinal products [5,7] or other medicinal products produced from cell culture [8]. Native trypsin from bovine or porcine pancreas is widely available for trypsinization, but concerns that such animal derived materials may introduce adventitious agents during manufacturing [9] (e.g. take the case of the paediatric vaccine Rotarix that was contaminated with porcine circovirus originating form batches of porcine trypsin, [10]) prompted the development of recombinant versions of the enzyme [11].
Impurity clearance
The clearance of nucleic acids from process streams arising from mammalian cell culture provides a good illustration of the class of applications of enzymes to DSP. In the manufacturing of adenovirus [12,13], lentivirus [14], influenza virus [15] and virus-like particles [16], for example, treatment with DNases (e.g. Benzonase® and Pulmozyme®) and RNases is usually performed prior to or after clarification to reduce cell nucleic acid content. This step not only improves purity, hence safety of the viral product [13], but also reduces the high viscosity associated with DNA and agglomeration of viral particles, which is usually induced by adhesion of nucleic acids [17]. In some cases, nuclease treatment can be carried out at the end of the process, after chromatographic purification [14, 18]. Nucleases can also be used to digest plasmid DNA vectors if a transient transfection strategy is used for viral particle production [19,20].
RNase is popular nuclease in the downstream processing of plasmid DNA due to its excellent ability to remove RNA from the producer E. coli cells [21,22]. While the benefits of RNase clearance to the performance of subsequent chromatographic operations are well known to those who have worked in pDNA purification, regulatory concerns with the animal origin of the benchmark RNAse A (bovine pancreas) have resulted in the banning of the enzyme from the process scene. An obvious way to circumvent this problem would be to use recombinant RNAse [23] or native microbial RNases [24], but surprisingly this solution has not been adopted by manufacturers. Exonucleases have also proved valuable in the lab scale purification of plasmid DNA due to their ability to digest linear DNA fragments that originate from the shearing/ degradation of genomic DNA or from plasmid linearization are [25].
Targeted structural modifications
In the first two categories of applications described, enzymes are used for they ability to break apart biomacromolecules from producer cells. The third category, however, includes more sophisticated applications, where a very precise alteration of the structure of a target molecule or impurity is required. Take the case of affinity tagremoval, for example [26,27]. These exogenous amino acid sequences have a high affinity for a specific ligand and are usually added via genetic design to proteins to facilitate production and purification. The use of His-tags [28] and of the octapeptides FLAG [29] and Streptag II [30], for example, make it possible to purify the corresponding tag-protein fusions by affinity chromatography with immobilized nickel, antibody and streptavidin matrices, respectively [26]. Other tags are used with the goal of improving fusion solubility like Glutathione S-transferase (GST), Maltose binding protein (MBP) and Elastin-like polypeptides (ELP). Purification is also facilitated with these tags due to their ability to bind to specific matrices (e.g. glutathione–Sepharose for GST, amylose for MBP) or to undergo temperature-induced aggregation (ELPs) [26]. Whichever is the reason for introducing a tag, in most applications the removal of the tag is a requirement. Here, proteolytic enzymes are used to precisely clip away the tag from the target protein by hydrolyzing the peptidic bond at a specific given location within the fusion. Exoproteases (e.g. carboxypeptidase A and B, DAPase) or endoproteases (e.g. tobacco etch virus protease, Factor Xa, thrombin, enteropeptidase and rhinovirus 3C proteases) are selected to remove tags, depending on the exact genetic design of the fusion and location of the tag [27].
Proteases have also played an important role in a number of processes used to produce recombinant insulin [31-33] and antibody fragments [34-38]. In the first case, the key role of proteases is to enzymatically transform precursor proinsulin molecules produced intracellularly by E. coli (via mixtures of trypsin and carboxypeptidase B) or extracellularly by S. cerevisiae (via trypsinmediated transpeptidation) into insulin [31,33]. In the second case, antibody fragments for therapeutic and diagnostic applications can be obtained by controlled proteolytic digestion of whole antibodies (mono- or polyclonal). For example, papain digestion yields two Fab and one Fc fragment [34-36], whereas the targeted attack of pepsin below the hinge region originates a bivalent F(ab’)2 fragment and a pFc’ fragment [37,38].
The glycan structure of proteins is modified in vivo via a set of specific enzymes. This suggests that proteins produced by a prokaryotic host, for example, can be glycosylated in vitro according to the glycan pattern that is more appropriate for the final biopharmaceutical application [39-41]. For example, the formation of the glycosidic bond between the polypeptide and chemically-synthesized glycan parts can be carried out by specialized enzymes like endoglycosidases (Endo-A [42], Endo-M [40] and Endo-F [43]) via a transglycosylation reaction [41]. This type of strategy could open-up the way for the synthesis of homogeneously glycosylated proteins with glycan side chains at natural or unnatural glycosylation sites.
HPrecise enzyme-catalyzed structural modifications have also targeted key impurities with the goal of facilitating their separation from the target molecule. Two notable examples can be found in the case of the very challenging separation of minicircle (MC) DNA vectors from miniplasmid (MP) impurities, two species very similar in terms of structure and size. In one case, in vitro restriction with an adequate enzyme is used to linearize MP and facilitate separation by gradient centrifugation [44]. In the second example, a nicking endonuclease modifies the topology of the MP from supercoiled to open circular, making it possible to subsequently use a hydrophobic interaction chromatography step that separates closed DNA molecules according to their isoform [45].
Considerations
While a few enzyme-assisted process steps have gained industry acceptance (e.g. the trypsin/ carboxypeptidase C excision of the C-peptide from proinsulin [32]), the fact that more of them have not entered into the biopharmaceutical downstream processing practice and rather remained stuck in the limbo of academic labs is somehow intriguing. This is even more puzzling if one considers the vast number of enzyme activities at our disposal that could be explored for purification purposes. Arguments against the use of enzyme preparations in the downstream processing of biopharmaceuticals typically point to: (i) a lack of cost effectiveness and commercial availability and (ii) hurdles associated with regulatory approval.
Although at first sight the lack of cost effectiveness/commercial availability argument is compelling, it is also evident that technologies are available nowadays that could be mobilized to produce process-scale amounts of enzymes at a reduced cost, as long as there is a clear demand from the biopharmaceutical industry for those products. After all, enzymes are produced at a cost low enough so that they can be incorporated into products like detergents (e.g. subtilisin), food additives (e.g. chymosin) and textile aids (e.g. pectinase, cellulase). The highest purity required for enzymes used in the manufacturing of pharmaceuticals is of course a major difference, but the fact that the use of enzymes could improve downstream processing by decreasing the number of process steps and associated labour, reducing the impact of impurities in chromatography operations or improving final product quality, should compensate for the higher cost of such enzymes. If one takes the example of protein A, it seems reasonable to assume that companies producing high-value products like biopharmaceuticals are able to accommodate the use of expensive mass separating agents.
Additionally, a final decision on whether to use or not an enzyme such only be taken on the basis of a professional cost analysis such as the one provided by modelling tools like SuperProDesigner (Intelligen Inc., Scotch Plains, NJ, USA), as exemplified for the case of insulin [33]. In this case study, the cost associated with ~2 kg of enzymes required to produce 1500 kg of insulin represented less than 1.5 % of the total costs of raw materials. This interesting example warns us that enzyme costs may not be as significant as one might anticipate.
Issues related to regulatory approval are also critical, as judged by reports of contamination [10] and by the concerns raised by the FDA and EMA with the animal origin of some of the most interesting enzymes used in recovery and purification (e.g. trypsin, pepsin, RNase) [9,47]. One obvious way to avoid the more stringent regulations in these cases is to resort to animal-component-free recombinant enzymes [11,23]. For example, recombinant trypsin is already available in the market [11] and there is no reason why other animal-derived enzymes cannot follow the same path. Clearly, the biopharmaceutical industry should be able to reap the benefits of enzyme activity and diversity, particularly in the downstream processing area, in a more systematic and rational way.
References
- Andrews BA, Asenjo JA. Enzymatic lysis and disruption of microbial cells.Trends. Biotechnol.5, 273-277 (1987).
- Salazar O, Asenjo JA. Enzymatic lysis of microbial cells.Biotechnol. Lett. 29, 985-994 (2007).
- Middelberg APJ. Process-scale disruption of microorganisms.Biotechnol. Adv.13, 491-551 (1995).
- Vogels G, Kula MR. Combination of enzymatic and/or thermal pretreatment with mechanical cell disintegration.Chem. Eng. Sci.47, 123-131 (1992).
- Codinach M, Blanco M, Ortega I, et al. Design and validation of a consistent and reproducible manufacture process for the production of clinical-grade bone marrow-derived multipotentmesenchymal stromal cells. Cytotherapy. 18,1197-1208 (2016).
- Wu F, Reddy K, Nadeau I, Gilly J, Terpening D, et al. Optimization of a MRC-5 cell culture process for the production of a smallpox vaccine.Cytotechnology 49, 95-107 (2005).
- Kami D, Watakabe K, Yamazaki-Inoue M, et al. Large-scale cell production of stem cells for clinical application using the automated cell processing machine. BMC Biotechnology. 13, 102 (2013).
- Sinacore MS, Drapeau D, Adamson SR. Adaptation of mammalian cells to growth in serum-free media. Mol. Biotechnol. 15, 249-257 (2000).
- Berger CN, Le Donne P, Windemann H. Use of substances of animal origin in pharmaceutics and compliance with the TSE-risk guideline -- a market survey. Biologicals.33, 1-7 (2005).
- Baylis SA, Finsterbusch T, Bannert N, Blümel J, Mankertz A. Analysis of porcine circovirus type 1 detected in Rotarix vaccine. Vaccine. 29, 690-697(2011).
- Novozymes. rTrypsin. Product Data Sheet (2013).
- Kamen A, Henry O. Development and optimization of an adenovirus production process. J.Gene. Med.6, S184-192 (2004).
- Konz JO, Lee AL, Lewis JA, Sagar SL. Development of a purification process for adenovirus: controlling virus aggregation to improve the clearance of host cell DNA. Biotechnol. Prog.21, 466-472 (2005).
- Bandeira V, Peixoto C, Rodrigues AF, et al. Downstream processing of lentiviral vectors: Releasing bottlenecks. Hum. Gene.Ther. Meth. 23, 255-263 (2012).
- Weigel T, Solomaier T, Peuker A, et al. A flow-through chromatography process for influenza A and B virus purification.J. Virol. Methods. 207, 45-53 (2014).
- Steppert P, Burgstaller D, Klausberger M, et al. Purification of HIV-1 gag virus-like particles and separation of other extracellular particles. J. Chromatogr. A. 1455, 93-101 (2016).
- Zhang S, Thwin C, Wu Z, Cho T. Method for the production and purification of adenoviral vectors. US patent application 6194191 B1, Feb 27, 2001.
- Green AP, Huang JJ, Scott MO, et al. A new scalable method for the purification of recombinant adenovirus vectors.Hum. Gene Ther. 13, 1921-1934 (2002).
- Sastry L, Xu Y, Cooper R, Pollok K, Cornetta K. Evaluation of plasmid DNA removal from lentiviral vectors by Benzonase treatment. Hum. Gene Ther. 15, 221-226 (2004).
- Shaw A, Bischof A, Jasti A, et al. Using PulmozymeDNase treatment in lentiviral vector production. Hum. Gene.Ther. Meth. 23, 65-71 (2012).
- Varley DL, Hitchkock AG, Weiss AME, et al. Production of plasmid DNA for human gene therapy using modified alkaline cell lysis and expanded bed anion exchange chromatography.Bioseparation. 8, 209-217 (1998).
- Prazeres DMF, Schluep T, Cooney C. Preparative purification of supercoiled plasmid DNA using anion exchange chromatography. J. Chrom. A. 806, 31-45 (1998).
- Voss C, Lindau D, Flaschel E. Production of recombinant RNase Ba and its application in downstream processing of plasmid DNA for pharmaceutical use. Biotechnol. Prog. 22, 737-744 (2006).
- Hames EE, Demir T. Microbial ribonucleases (RNases): production and application potential. World. J. Microbiol. Biotechnol. 31, 1853-1862 (2015).
- Balagurumoorthy P, Adelstein SJ, Kassis AI. Method to eliminate linear DNA from mixture containing nicked-circular, supercoiled, and linear plasmid DNA.Anal. Biochem. 381, 172-174 (2008).
- Arnau J, Lauritzen C, Petersen GE, Pedersen J. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein.Expr.Purif. 48,1-13 (2006).
- Waugh S. An overview of enzymatic reagents for the removal of affinity tags. Protein.Expr. Purif. 80, 283-293 (2011).
- Hoffmann A, Roeder RG. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res.19,6337-6339 (1991).
- Knappik A, Pluckthun A. An improved affinity tag based on the FLAG peptide for the detection and purification of recombinant antibody fragments, Biotechniques 17, 754-761 (1994).
- Skerra A, Schmidt TGM. Use of the Strep-tag and streptavidin for detection and purification of recombinant proteins, Methods. Enzymol. 326, 271-304 (2000).
- Kehoe JA. The story of biosynthetic human insulin. In: Frontiers in Bioprocessing, CRC Press, Boca Raton, 1990, pp. 45-50.
- Walsh G. Therapeutic insulins and their large-scale manufacture. Appl. Microbiol. Biotechnol. 67, 151-159 (2005).
- Petrides D, Sapidou E, Calandranis J. Computer-aided process analysis and economic evaluation for biosynthetic human insulin production-a case study. Biotechnol. Bioeng. 48, 529-541 (1995).
- Adamczyk M, Gebler JC, Wu J. Papain digestion of different mouse IgG subclasses as studied by electrospray mass spectrometry. J. Immunol. Meth. 237, 95-104 (2000).
- Cresswell C, Newcombe AR, Davies S, Macpherson I, Nelson P, O’Donovan K, Francis R. Optimal conditions for the papain digestion of polyclonal ovineIgG for the production of biotherapeutic Fab fragments. Biotechnol.Appl. Biochem. 42, 163-168 (2005).
- Thillaivinayagalingam P, O'Donovan K, Newcombe AR, Keshavarz-Moore E. Characterisation of an industrial affinity process used in the manufacturing of digoxin-specific polyclonal Fab fragments.J. Chromatogr. B. 848, 88-96 (2007).
- Kumpalume P, Boushaba R, Jones RGA, Slater NKH. FacileF(ab’)2 manufacturing: Strategies for the production of snake antivenoms. Trans. IchemE. 80, 80-97 (2002).
- Boushaba R, Kumpalume P, Slater NKH. Kinetics of whole serum and prepurifiedIgG digestion by pepsin for F(ab′)2 manufacture. Biotechnol. Prog. 19, 1176-1182 (2003).
- Meynial-Salles I, Combes D. In vitro glycosylation of proteins: an enzymatic approach. J. Biotechnol. 46, 1-14 (1996).
- Yamamoto K. Chemo-enzymatic synthesis of bioactive glycopeptide using microbial endoglycosidase. J. Biosci. Bioeng.92, 493-501 (2001).
- Khmelnitsky YL. Current strategies for in vitro protein glycosylation.J. Mol. Catal. B: Enzymatic. 31, 73-81 (2004).
- Takegawa K, Tabuchi M, Yamaguchi S, et al. Synthesis of neoglycoproteins using oligosaccharide-transfer activity with endo-b-N-acetylglucosaminidase. J. Biol. Chem. 270, 3094-3099 (1995).
- Yamamoto K, Takegawa K. Transglycosylation activity of endoglycosidases and its application. Trends.Glycosci. Glyc. 9, 339-354 (1997).
- Darquet A, RangaraR, Kreiss P, et al. Minicircle: an improved DNA molecule for in vitro and in vivo gene transfer. Gene.Ther. 6, 209-218 (1999).
- Alves CPA, Simcíková M, Brito L, Monteiro GA, Prazeres DMF. Development of a nicking endonuclease-assisted method for the purification of minicircles.J. Chrom. A. 1443, 136-44 (2016).
- Committee for Medicinal Products for Human use (CHMP). Guideline on the use of porcine trypsin used in the 4 manufacture of human biological medicinal products. EMA/CHMP/BWP/814397/2011