Short Communication - Imaging in Medicine (2016) Volume 8, Issue 4
Quantitative analysis of threedimensional branching systems from X-ray computed microtomography data
Adriana L McKinney and Tamas Varga*Pacific Northwest National Laboratory, Richland, WA, USA
- *Corresponding Author:
- Tamas Varga
Pacific Northwest National Laboratory
Richland, WA, USA
Tel : (509) 371-6042
Fax: (509) 371-7866
Email: tamas.varga@pnnl.gov
Abstract
Branching structures such as lungs, blood vessels and plant roots play a critical role in life. Growth, structure, and function of these branching structures have an immense effect on our lives. Therefore, quantitative size information on such structures in their native environment is invaluable for studying their growth and the effect of the environment on them. X-ray computed tomography (XCT) has been an effective tool for in situ imaging and analysis of branching structures. We developed a costless tool that approximates the surface and volume of branching structures. Our methodology of noninvasive imaging, segmentation and extraction of quantitative information is demonstrated through the analysis of a plant root in its soil medium from 3D tomography data. XCT data collected on a grass specimen was used to visualize its root structure. A suite of open-source software was employed to segment the root from the soil and determine its isosurface, which was used to calculate its volume and surface. This methodology of processing 3D data is applicable to other branching structures even when the structure of interest is of similar X-ray attenuation to its environment and difficulties arise with sample segmentation.
Background
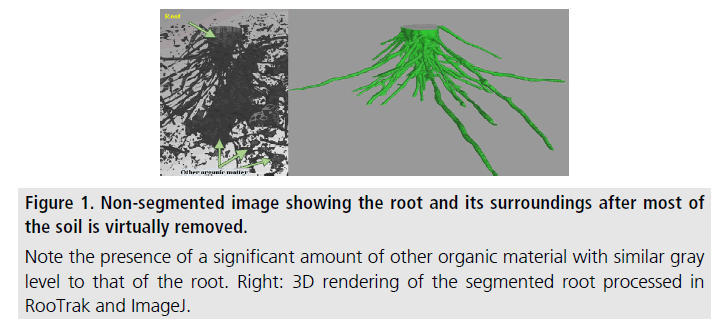
Numerous forms of branching structures are present in nature and play critical roles in life. Examples of such biological structures include; bronchial tissue in lungs, human and animal blood vessels, vein structures in leaves, and plant roots. In the present study, we look at plant roots that represent an "invisible" part of plant biology since soil makes it difficult to image roots noninvasively. However, the described methodology of imaging and quantitative analysis is applicable to other branching structures as long as threedimensional imaging data can be obtained on them. Studying root growth and interaction within the soil environment is essential to understanding root/plant growth and nutrient cycling, which in turn affect farming, forestation, food security, and climate. X-ray computed tomography (XCT) has proven to be an effective tool for noninvasive imaging of plant root samples in the soil. Following segmentation of the root from that of the surrounding soil in the tomography data, i.e., virtually isolating the root image from everything else around it, quantitative information, such as root surface area and volume can be extracted from the data. The challenges associated with in-soil imaging of plant roots include the similarities in the X-ray attenuation values between root and soil, in part caused by water and organic matter, and variations in X-ray attenuation properties of the root material. Available opensource computational tools, such as RooTrak [1,2] used in this work, are able to address these issues and successfully segment the root by tracking it through each image slice. Following segmentation, the isosurface of the segmented root system was represented by a triangular mesh, using an algorithm implemented in another open-source program, ImageJ. Finally, our code imeshJ was employed to calculate the root surface area and volume from the isosurface.
Method & Results
The specimen, two stems of the native grass Prairie drop seed (Sporobolus heterolepis) in original soil, was placed in a 1 inch diameter tubular plant holder, with no additional preparation. XCT imaging was performed using an X-Tek/Metris XTH 320/225 kV system (Nikon Metrology, Belmont, CA). Data was collected at 85 kV and 190 μA X-ray power. The sample was rotated continuously during the scan with momentary stops to collect frames for each projection (shuttling mode) to minimize ring artifacts. 3142 projections were collected over 360 degrees with 1 s exposure time per frame and 4 frames per projection. After acquisition the software CT Pro 3D (Metris XT 2.2, Nikon Metrology, Belmont, CA) was used for reconstruction. The reconstructed data voxel size was approximately 31 × 31 × 31 μ. The images were brightened to increase contrast between root and soil values. The reconstructed volume file was used to create a top view image stack using the open-source image-processing program ImageJ 1.6 [3]. The reconstructed data clearly depicted that the root and other components, most likely organic matter, had very similar X-ray attenuation factors. This resulted in little to no grey-scale contrast in the images; showing the need for segmentation through RooTrak (Figure 1). RooTrak tracks continuity in roots as they branch out through the soil, and isolates the roots from surrounding material to produce a stack of images that contain only the root. To begin tracking, a seed point was selected inside each of the pertinent root sections visible in the top slice of the volume data, parameters “smoothness” and “similarity” were set to 0.3 and 0.8, respectively, then the tracking function of the software was initiated. After opening the output images in ImageJ, it was evident that RooTrak successfully segmented the selected ∼200 slices of volume data which corresponded to a length of 6.2 mm. Next, ImageJ was used to generate a triangular isosurface mesh of the 3D volume (approximating the surface of the isolated root) from the output produced by RooTrak. The mesh was saved in the binary stereolithography (STL) format, and the imeshJ script, developed by our group, was used to compute the quantitative measures of the isosurface. The output values of imeshJ are surface area in mm2 and volume in mm3. For the reconstructed volume in this study, the calculated surface area was 344.98 mm2 and the volume was 49.54 mm3. The accuracy of the calculated surface area is dependent on the assumption that the isosurface produced by ImageJ is a reasonable approximation to the actual surface of the root. This assumption is reasonable for large root segments, but may prove lesser so when roots with dimensions comparable to the voxel size are imaged. For such small-diameter roots, the voxel representation becomes less accurate. The accuracy of the calculated volume figure is dependent on the above assumption, but to a smaller degree because smaller-diameter roots contribute proportionally less to the volume of the root than the surface area of the root.
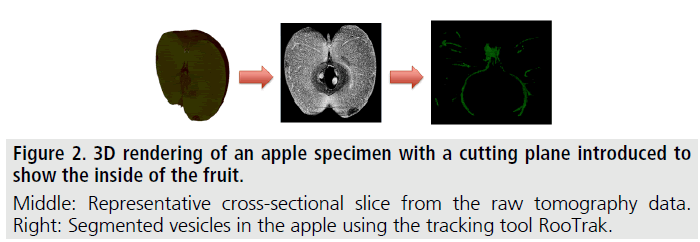
XCT and therefore extracting and evaluating branching structures is readily transferrable to other biological branching structures, such as, for instance, bronchial tissue and blood vessels in lungs [4] and the vesicular system in fruits. Figure 2 shows XCT images of an apple, where the fruit’s vesicular system was successfully tracked and segmented by the authors using the above described methodology.
Conclusion
A combination of X-ray computed tomography and open-source programs proved to be a powerful tool to noninvasively image plant root samples, segment root data, and extract quantitative information from the 3D data. Our ability to visualize and measure features depends on scan resolution, and the capabilities of the RooTrak software. In this model, scan resolution was sufficient to capture a vast majority of the features in the sample with ideal contrast enabling RooTrak to successfully segment the majority of the root. This methodology of processing 3D data should be applicable to other material/sample systems where there is continuity between components of similar X-ray attenuation and where difficulties arise with sample segmentation.
Acknowledgement
This work was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory.
References
- Mairhofer S, Zappala S, Tracy SR, et al. RooTrak: Automated recovery of three-dimensional plant root architecture in soil from x-ray microcomputed tomography images using visual tracking. Plant. Physiol. 158, 561-569 (2012).
- Mairhofer S, Zappala S, Tracy S, et al. Recovering complete plant root system architectures from soil via X-ray mu-Computed Tomography. Plant. Methods. 9, 8 (2013).
- ImageJ: Image Processing and analysis in Java. (2014).
- Scott AE, Vasilescu DM, Seal KAD, et al. Three dimensional imaging of paraffin embedded human lung tissue samples by micro-computed tomography. Plos. One. 10 (2015).