Research Article - Pharmaceutical Bioprocessing (2025) Volume 13, Issue 1
Efficiency of Baculovirus Clearance Methods from Biopharmaceuticals
Mohammad Reza Sadrzadeh, Mohsen Dehghani, Parisa Hashemi, Ehsan Seyedjafari*
Department of Biotechnology, University of Tehran College of Science, Tehran, Iran
- *Corresponding Author:
- Ehsan Seyedjafari
Department of Biotechnology, University of Tehran College of Science, Tehran, Iran
E-mail: seyedjafari@ut.ac.ir
Received: 10-Jan-2024, Manuscript No. FMPB-24-124859; Editor assigned: 12-Jan-2024, PreQC No. FMPB-24-124859 (PQ); Reviewed: 26-Jan-2024, QC No. FMPB-24-124859; Revised: 13- Feb-2025, Manuscript No. FMPB-24-124859 (R); Published: 20-Feb-2025, DOI: 10.37532/2048-9145.2025.13(1).258-267
Abstract
Viral clearance study is necessary to assure biological products to be free of viruses either introduced into the product via biological raw materials or adventitiously, and for conforming with continuously stricter rules of health authorities. As insect cell–baculovirus expression system has been considered as a powerful workhorse for recombinant protein production and is being harnessed in industrial manufacturing of several of biopharmaceuticals including HPV vaccine, Influenza vaccine and a prostate cancer immunotherapy, assurance of baculovirus removal from the final product is necessary as a relevant virus whose presence in the bioprocess is known. Hence, investigating ways of removal or inactivation of baculovirus for their incorporation in the manufacturing process is a fundamental step toward reaching that goal. Removal of baculovirus by anion exchange and mixed-modal chromatography and also by filtration through virus filters and its inactivation by UV-C, 2-ME, H2O2, low pH, high pH and pasteurization are considered in this research. Furthermore, as viral clearance study is process dependent, removal and inactivation of baculovirus has also been investigated in a human papillomavirus vaccine downstream process consisting of a hypotonic lysis of harvested cells, chromatography steps, virus filtration and ultrafiltration. Removal and inactivation of baculovirus is assessed by qPCR and TCID50 respectively. pasteurization treatment produced the highest level of inactivation with 5.87 LRV followed by H2O2, low pH, Triton X-100, 2-ME, UV-C with 5.58 4.33, 3.62, 3.58 and 2.58 LRVs respectively. High pH resulted in a negligible inactivation. DMAE chromatography, Viresolve virus filter and CHTII chromatography resulted in 4.90, 4.64 and 3.90 LRV respectively.
Keywords
Viral clearance study • Baculovirus • Inactivation • Log reduction value • Bioprocess • Virus titration
Introduction
One of the most important aspect of biopharmaceuticals safety is to ensure the product is free of viruses and other transmissible agents. Throughout history, many cases of viral contamination of biopharmaceuticals has occurred with devastating effects including paralysis, HIV infection and even death with the peak being in the late 20th century [1]. This hard gained knowledge has led to the development of prophylactic measures taken by the manufacturers and legislated by health authorities. According to ICH guidelines, the constitution of biopharmaceuticals production, any manufacturer of biological products, especially those using animal-derived material in their production process has to take actions to ensure no contamination of the product by viruses and other transmissible agents has occurred and if this contamination is inevitable, removal or inactivation of the contaminant is guaranteed via downstream purification processes. This level of assurance is demonstrated by state of the art viral clearance studies [2].
In viral clearance studies, the ability of the process to remove or inactivate viruses is assessed by deliberately spiking specified amount of different viruses individually, into the inputs of downstream process unit operations and titrating the amount of virus present in the output of the corresponding unit operations. The level of reduction in virus titer is then calculated and is considered a quantitative measure of the ability of that unit operation to remove or inactivate virus. For this purpose, different viruses should be chosen with different levels of physicochemical resistance covering both RNA/DNA, single stranded/double stranded viruses. Viruses that are known to be present or contaminate the process are called relevant viruses.
Baculovirus is an eveloped rod-shaped virus with dimensions 30-60 nm × 250-300 nm, containing a supercoiled dsDNA, 80-180 kbp in size [3]. Physicochemical resistance of this virus assessed in low to medium category. Occlusion bodies are stable form of virus with increased resistance.
Baculovirus is nowadays considered the relevant virus of recombinant protein production processes using the popular insect cell-baculovirus expression system. As of today three commercial products, HPV vaccine, Influenza vaccine and a cell-based immunotherapy use baculovirus in their manufacturing process. As information on the removal and/or inactivation of baculovirus and the efficiency of unit operations to do so is limited in academic and industrial literature, this research is dedicated to investigate ways to remove and inactivate this virus. Common viral removal/incactivation methods are extracted from the literature [4].
Since any viral clearance study is inevitably to some extent process dependent, as a case study, the manufacturing process of bivalent HPV vaccine, cervarix is extracted from the patent of the manufacturer, GlaxoSmithKline and a viral clearance study is performed for the extracted process in a scaled-down purification process [5].
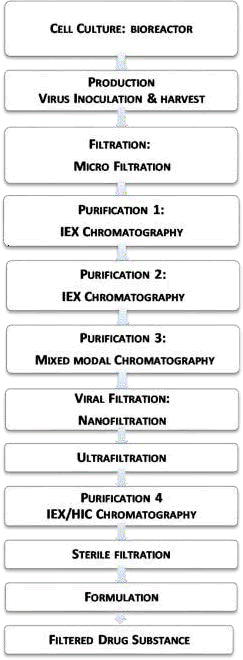
Production process of HPV vaccine according to GSK is as shown in the flowchart of Figure 1.
Figure 1. Flowchart of HPV type 16 L1 protein proposed by GlaxoSmithKleine: Purification 1 is DMAE column chromatography, Purification 2 is TMAE column chromatography, Purification 3 is CHTII column chromatography and Purification 4 is DEAE column chromatography.
Other unit operations not present in GSK process including treatment with H2O2, UV- C, 2-ME, low pH, high pH and pasteurization have been assessed for their ability to baculovirus inactivation. Furthermore, as commercial virus filters differ in their level of virus removal, Viresolve virus filter has been assessed for virus removal in contrast to the GSK process which uses Planova virus filters.
Materials and Methods
Baculovirus propagation
Sf9 cell culture is performed according to the state of the art. Sf9 cell back (purchased from Invitrogen) is thawed and passaged in 75 cm2 T-flask using Sf900II media (purchased from Thermofisher) supplemented with 10% FBS and incubated in 27°C incubator. After reaching 90% confluency (24 h), cells are transferred to a spinner flask for cell suspension adaptation. Cells are then cultured in a shake flask with initial cell density of 5 × 105 cells/ml. when cell density of 2 × 106 is reached, cells are infected with MOI=0.1 of recombinant HPV L1 baculovirus. After 96 hours of incubation, baculovirus can be harvested in the supernatant of the cell culture by centrifugation. Two different stocks of baculovirus were prepared with titers 109 and 3 × 108.
Production of HPV 16 L1
High five insect cell culture is performed according to the state of the art [6]. Hi5 cell bank (purchased from Invitrogen) is thawed and passaged in 75 cm2 T-flasks using ExpressFive media (purchased from Thermofisher) supplemented with 10% FBS and incubated in 27°C incubator. After reaching 90% confluency (48 h), cells are further passaged. Then cells are transferred for cell suspension adaptation. Cells are then cultured in a shake flask l and incubated until cell density of 3 × 106 is reached. After several passages, several shake flasks containing 7 × 109 cells are pooled and transferred to a WAVE bioreactor diluting the cells up to 5 L with media. Cells are infected with MOI=1.5 of recombinant HPV L1 baculovirus. After 72 hours of incubation, cells are harvested and lysed [7]. Lysed and clarified cells are applied to 3 chromatography columns and nanofiltered by Viresolve virus filter.
DMAE chromatography
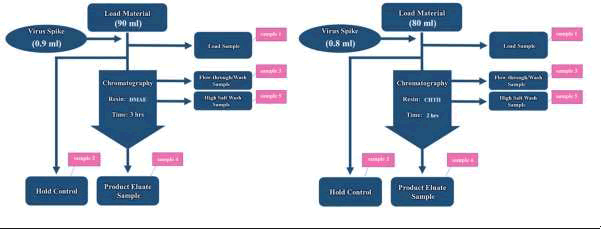
Anion-exchange chromatography in Bind and Elute (B and E) mode is performed in a scaled- down model at room temperature (25°C) to assess its virus removal ability. 10 ml of DMAE resin (purchased from GE) is packed into XK 16/20 column (GE). 90 ml of Hi5 cell supernatant was lysed and spiked with 0.9 ml of a stock baculovirus with titer of 109 determined with qPCR (A low spike ratio (1%) was selected to ensure that the chemical characteristics of the challenge solution remained unaltered). 5 ml of the sample is held as control. In each step a 3 ml-sample is taken as shown in Figure 2. Flow in all steps is 1 ml/min. Column is equilibrated with equilibration buffer (20 mM Tris, pH 7.9). Spiked sample is applied to the column. Flow through output is collected. Column is then washed with washing buffer (20 mM Tris, pH 7.9). To demonstrate typical elution profiles, Elution of HPV16 L1 protein is performed with 5 column volume of elution buffer (20 mM Tris, 200mM NaCl, 4% 2-ME, pH 7.9) until protein peak is fully recovered.
Figure 2. Schematic process of virus spiking on DMAE and CHTII column chromatography.
CHTII chromatography
Mixed-modal chromatography in B and E mode is performed in a scaled-down model at room temperature (25°C) to assess its virus removal ability. 5 ml prepacked CHTII column (purchased from Bio-Rad) is used for this purpose. 80 ml of TMAE eluate is spiked with 0.8 ml of a stock baculovirus with titer of 109 determined with qPCR. 5 ml of the sample is held as control. In each step a 3 ml-sample is taken as shown in Figure 2. Flow in all steps is 1 ml/min. Column is equilibrated with equilibration buffer (20 mM Tris, 50 mM NaCl, pH 8.5). Spiked sample is applied to the column. Flow through output is collected. Column is then washed with washing buffer (20 mM Tris, 50 mM NaCl, pH 8.5). Elution of HPV16 L1 protein is performed with 5 column volume of elution buffer (NaH2PO4 100 mM, NaCl 30 mM, 4% 2-ME, pH 6) until protein peak is fully recovered.
Virus filtration
0.5 ml baculovirus with titer 3 × 108 was added to 0.5 ml of CHTII elution buffer (NaH2PO4 100 mM, NaCl 30 mM, 4% 2-ME) at room temperature (25°C). Using a 1 ml insulin syringe, the sample was applied to Viresolve Pro Micro Device (purchased from Merck) with the minimum pressure possible.
Ultrafiltration/diafiltration
The viruses after each treatment were diafiltered 10X against PBS (pH=7.4) using Ultra-15 30 kDa Amicon (purchased from Merck) with 2500 RCF in Universal 320R centrifuge.
2-ME treatment
0.6 ml from the stock solution of baculovirus with titer 3 × 108 (determined by qPCR) was added to 5.4 ml of DMAE elution buffer containing 4% 2-ME. The prepared solution was incubated at room temperature and after 2 h, 4 h and 8 h, 2 ml of the solution was taken and immediately diafiltered.
H2O2 treatment
200 μL from the stock solution of baculovirus with titer 3 × 108 (determined by qPCR) was added to 5.4 ml of DMAE elution buffer containing 3% H2O2 (from stock solution of 30%). The prepared solution was incubated at room temperature for 8 h. The solution was immediately diafiltered.
Pasteurization
To 1.8 ml of DMAE elution buffer was added 200 μL from the stock solution of baculovirus with titer 3 × 108 (determined by qPCR). The prepared solution was incubated at 60°C for 9 h. No diafiltration was performed.
Low pH treatment
To 1.8 ml of DMAE elution buffer was added 200 μL from the stock solution of baculovirus with titer 3 × 108 (determined by qPCR). The pH of the solution was adjusted to pH=4 by adding appropriate amount of HCl 0.1M. The prepared solution was incubated at room temperature for 7 h. The solution was immediately diafiltered.
High pH treatment
To 1.8 ml of DMAE elution buffer was added 200 μL from the stock solution of baculovirus with titer 3 × 108 (determined by qPCR). The pH of the solution was adjusted to pH=10 by adding appropriate amount of NaOH 0.2 M. The prepared solution was incubated at room temperature for 7 h. The solution was immediately diafiltered.
Triton X-100 treatment
To 1.8 ml of DMAE elution buffer was added 200 μL from the stock solution of baculovirus with titer 3 × 108 (determined by qPCR). 1% v/v Triton X-100 was then added to the solution. The prepared solution was incubated at room temperature for 30 min. The solution was immediately diafiltered.
TCID50
The samples of inactivation treatments titers were based on the 50% Tissue Culture Infectious Dose (TCID50) endpoint in Sf9 cells according to Roldao, et al. [8]. Tenfold, serially diluted samples were inoculated into a 96-well microplate, seeded with Sf9 cells with a density of 2 × 105 cells/ml, in eight replicate (10 μL per well) and incubated for 7 days in 27°C incubator placed in a box containing a dampened sterile gauze. 20 μL of each well was then transferred into a second microplate, seeded with Sf9 cells with a density of 2.5 × 105 cells/ml and incubated for 4 days. CPE incidence is then investigated microscopically. The solution titer was determined by Spearman-Karber method [9]. The baculovirus stock used in these studies contained 5.623 × 108 TCID50/mL (3 × 108 by qPCR).
qPCR for viral nucleic acid extraction, DynaBio Viral Nucleic Acid Extraction Mini Kit (Takapuzist, Cat. No. 10140114) was used. qPCR was performed on the Corbett Rotor-Gene 6000 (Rotor-Gene software version 1.7.78) using Maxima SYBR Green qPCR master mix (Thermo Scientific, Cat. No. K0222) and primers (Fwd: GGAGCTGATGTACGAAAACGAT; Rev: GGTCGTGCAGCATATTGTTTAG) against gp64 coding sequence, the main glycoprotein of Autographa Californica Nucleopolyhedrovirus with NCBI accession number NC_001623.1. BaculoQuant kit (purchased from Oxford Technologies) standard virus stock was used to prepare an internal standard for qPCR titration.
LRV calculation
LRV calculation is performed according to ICH Q5A (2) using the formula below: LRV=(log10(V1)+log10(T1))- (log10(V2)+log10(T2)).
Where V1 and T1 are the volume and titer of the input starting material and V2 and T2 are the volume and titer of the output treated material respectively.
Confidence interval of LRV is calculated using the formula below:
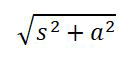
s and a are the standard deviation of titer determination assay before and after performing the virus removal or inactivation procedure.
Results
TCID50
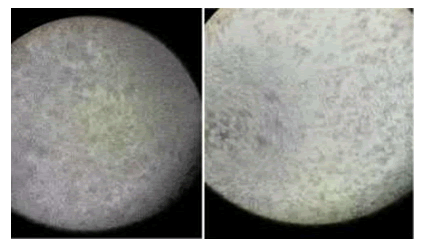
CPE in Sf9 cells in TCID50 assay is recognized by inhibition of cell growth and increase in cell size.
DMAE chromatography
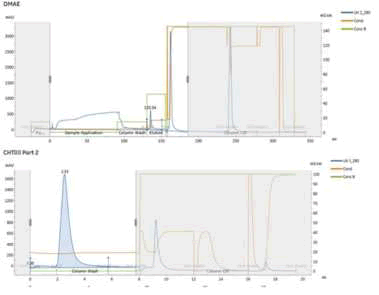
DMAE chromatogram (Figure 5) showed the peaks like an ordinary purification process without spiking.
Eluate and flow through samples are compared to the titer of the input sample loaded onto the column to assess both B and E and FTH modes of the column in virus removal abilities.
Reduction levels are summarized in Table 1.
Most of the viruses were recovered in the FTH fraction and eluate showed the highest LRV. Virus recovery was very low. Reduction levels are summarized in Table 2.
|
|
Units |
DMAE column |
|
Initial load sample initial count |
qPCR/ml |
107 |
|
Final load sample final count |
TCID50/ml |
6.76 × 105-107 (0%-93% loss |
|
Final eluate sample final |
qPCR/ml |
1.93 × 102 |
|
Final eluate sample final count rep 2 |
qPCR/ml |
1.53 × 102 |
|
Flow through sample final |
qPCR/ml |
1.01 × 106 |
|
Flow through sample final count rep 2 |
qPCR/ml |
8.99 × 105 |
|
Load volume |
ml |
82 |
|
Eluate volume |
ml |
60 |
|
FTH volume |
ml |
182 |
|
Eluate LRV |
LRV |
4.90 ± 0.14 |
|
FTH LRV |
LRV |
0.67 ± 0.07 |
|
Virus recovery count (mass balance) |
qPCR/ml |
1.84 × 108 (22% recovery) |
Table 1. DMAE column baculovirus clearance results.
|
|
Units |
CHTII column |
|
Initial load sample initial count |
qPCR/ml |
107 |
|
Final load sample final count (hold control) |
TCID50/ml |
6.76 × 105–107 (0%-93% loss of infectivity based on 95% CI) |
|
Final eluate sample final count rep 1 |
qPCR/ml |
4.36 × 103 |
|
Final eluate sample final count rep 2 |
qPCR/ml |
2.88 × 103 |
|
Flow through sample final count rep 1 |
qPCR/ml |
8.34 × 105 |
|
Flow through sample final count rep 2 |
qPCR/ml |
7.43 × 105 |
|
Load volume |
ml |
72 |
|
Eluate volume |
ml |
25 |
|
FTH volume |
ml |
122 |
|
Eluate LRV (based on mean titer) |
LRV |
3.91 ± 0.25 |
|
FTH LRV (based on mean titer) |
LRV |
0.87 ± 0.25 |
|
Virus recovery count (mass balance) |
qPCR/ml |
9.55 × 107 (13% recovery) |
Table 2. CHTII column baculovirus clearance results.
CHTII chromatography
CHTII chromatogram (Figure 3) showed the peaks like an ordinary purification process without spiking and HPV 16 L1 was eluted with elution step.
Eluate and flow through samples are compared to the titer of the input sample loaded onto the column to assess both B and E and FTH modes of the column in virus removal abilities.
Reduction levels are summarized in Table 1.
Most of the viruses were recovered in the FTH fraction and eluate showed the highest LRV. Virus recovery was very low. Reduction levels are summarized in Table 2.
Virus filtration
Nearly all the volume filtered was recovered in the permeate of the filter. Permeate was analyzed with qPCR. Virus reduction levels are summarized in Table 3.
Viral inactivation treatments 2-ME treatment
2-ME treatment: 2-ME virus inactivation results are summarized in Table 4.
Figure 3. Image of control non-infected wells in baculovirus TCID50 assay. right: Image of baculovirus infected wells showing CPE in TCID50 assay. CPE is in the form of cell inhibition and cell size increase. Images are recorded by a camera from an inverted microscope.
|
|
Units |
Virus filter removal results |
|
Initial challenge sample initial count |
qPCR/ml |
1.5 × 108 |
|
Filtrate sample final count |
qPCR/ml |
3.4 × 103 |
|
Volume filtered |
ml |
1 ml |
|
Filtrate LRV |
LRV |
4.64 ± 0.14 |
Table 3. Viresolve virus filter baculovirus clearance results.
|
|
Units |
2-ME inactivation results |
|
Sample initial count |
TCID50/ml |
5.623 × 108 (± 0.490 Log TCID50) |
|
2 h treatment sample count |
TCID50/ml |
5.623 × 107 (± 0.321 Log TCID50) |
|
4 h treatment sample count |
TCID50/ml |
3.16 × 107 (± 0.347 Log TCID50) |
|
8 h treatment sample count |
TCID50/ml |
5.62 × 104 (± 0.347 Log TCID50) |
|
Volume treated (each condition) |
ml |
2 |
|
2 h treatment LRV |
LRV |
0.583 ± 0.586 |
|
4 h treatment LRV |
LRV |
0.833 ± 0.600 |
|
8 h treatment LRV |
LRV |
3.583 ± 0.600 |
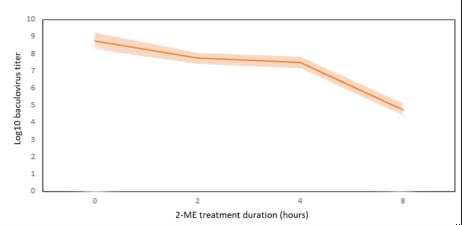
Table 4. 2-ME treatment baculovirus inactivation results after 2, 4 and 8 hours of incubation.
Kinetic of baculovirus inactivation with respect to time is illustrated in Figure 4. As inactivation speed was increased after 4 hours, investigation of treatment duration more than 8 hours shouldbe investigated (Figure 5).
Figure 4. kinetic of baculovirus inactivation by 2-ME treatment.
Figure 5. Full chromatogram of DMAE column chromatography in HPV 16 L1 protein purification (Top). Full chromatogram of CHTII column chromatography in HPV 16 L1 protein purification (Bottom).
Other treatments
Pasteurization treatment produced the highest level of inactivation followed by H2O2, low pH, 2-ME and UV-C. High pH resulted in a negligible inactivation.
Results of virus inactivation by H2O2, low pH, high pH, UV-C and pasteurization are summarized in Table 5.
|
|
Units |
|
|
Sample initial count |
TCID50/ml |
5.623 × 108 (± 0.490 Log TCID50) |
|
Volume treated |
ml |
2 |
|
H2O2 8 h treatment sample count |
TCID50/ml |
5.623 × 102 (± 0.321 Log TCID50) |
|
H2O2 8 h treatment LRV |
LRV |
5.583 ± 0.586 |
|
Low pH 7 h treatment sample count |
TCID50/ml |
104 (± 0.370 Log TCID50) |
|
Low pH 7 h treatment LRV |
LRV |
4.333 ± 0.614 |
|
High pH 7 h treatment sample count |
TCID50/ml |
2.371 × 107 (± 0.404 Log TCID50) |
|
High pH 7 h treatment LRV |
LRV |
0.958 ± 0.635 |
|
UV-C 30 min treatment sample count |
TCID50/ml |
5.623 × 105 (± 0.321 Log TCID50) |
|
UV-C 30 min treatment LRV |
LRV |
2.583 ± 0.586 |
|
Pasteurization 9 h treatment sample count |
TCID50/ml |
7.499 × 102 (± 0.404 Log TCID50) |
|
Pasteurization 9 h treatment LRV |
LRV |
5.875 ± 0.635 |
|
Triton X-100 30 min treatment sample count |
TCID50/ml |
1.334 × 105 (± 0.359 Log TCID50) |
|
Triton X-100 30 min treatment LRV |
LRV |
3.625 ± 0.607 |
Table 5. H2O2, low pH, high pH, UV-C and pasteurization, Triton X-100 baculovirus inactivation results.
Discussion
Baculovirus expression system is being massively used by academic and industrial section especially for recombinant protein production. In addition to curretly approved products, several other vaccines including diabetes and hepatitis E are being developed using BEVS technology.
As a successful case of BEVS technology, HPV vaccine by GSK can be considered. HPV vaccine has been introduced into the market since 2006 for prevention of cervix cancer and is considered an important vaccine by WHO, recomended to be incorporated into routine vaccination programs. As of today 5 HPV vaccines exist: Cervarix, Papiloguard, Cecolin, Gardasil and Gardasil 9. Several other manufacturers in India and China have speculated to reach their own HPV vaccine into the market by 2021 as the demand will be 100 million doses in the next 10 years [10].
Hence, investigation of viral clearance of baculovirus as a relevant virus in BEVS technology is of utmost importance for the safety of the final vaccine for the manufacturers and health authorities and this study was carried out to assess various orthagonal methods and treatments for removal and incativation of baculovirus with a case study for the assessment of viral clearance in the production of insect cell- derived HPV-16 L1 VLPs. For the production of HPV-16 L1 VLPs, High Five cells were used with baculovirus expression vector system.
The purification of HPV-16 L1 VLPs was performed according to Figure 1. Out of the unit operations of this process, chromatography and virus filter was assessed as potentially capable of removing the virus. As DMAE, TMAE and DEAE are all anion exchange chromatography columns performed in B and E mode the mechanism of removal for the three of them are the same and only DMAE was assessed. DMAE is prefered bacause it is the first column and in the real process it is probably in contact with the highest virus titer. In addition to DMAE, CHTII which is a mixed-modal chromatography column is also investigated as its mode of action for virus removal is different.
One of the challenges in viral clearance of baculovirus is having reliable titration methods to investigate its removal and inactivation. Especially for inactivation investigation, most of the protocols for TCID50 for live virus titration don’t actually work or are hard to interpret because recombinant baculovirus hardly produces CPE due to lack of occlusion bodies. Here we used a rather new protocol of TCID50 that uses a 7+4-day incubation period.
Throughout the purification process 2-ME is used for prevention of VLP self-assembly. As 2-ME is a reducing agent and baculovirus proteins especially gp64 the major protein of baculovirus envelope consists of many disulfide bonds, it can have a detrimental effect on virus activity.
In addition to the main steps and treatments already in the purification process, other treatments effects were also evaluated to be used as reference for future products using BEVS technology with different purification processes.
Each procedure was evaluated to determine a reduction factor for viral clearance. To minimize cytotoxic effect of the test reagents in TCID50 assay, after treating the virus with reagents, the treated virus was diafiltered against PBS buffer to remove the reagent and therefore, no cytotoxicity study was performed. In other studies, performed in the literature, the reagents are usually highly cytotoxic for the indicator cells of titration assays and the sample are diluted (up to 1:400) to the point where no nonspecific cytotoxicity is seen after treatment for carrying out the assays. This dilution may cause the sample to go below the limit of detection. So the ultrafiltration/ diafiltration step can be an ideal substitute to minimize cytotoxicity.
Reduction factors by DMAE, CHTII and Viresolve virus filter as removal steps were 4.90, 3.91 and 4.64 respectively. LRV showed for Viresolve virus filter was quite in agreement with other studies for other viruses which caused 4.8- 5.7 LRV for bacteriophage PP7 [11]. Reduction factors by 2-ME increased from 0.583 in 2h treatment to 0.83 and 3.58 in 4 h and 8 h treatments respectively. In literature there is no report of 2-ME for viral inactivation. However, the obtained results are in agreement with another study, investigating DTT as a reducing agent for virus inactivation which resulted in between 3.5-5.1 LRV for different viruses [12]. The kinetics of viral inactivation by 2-ME can be seen in Figure 4.
In chromatography removal steps, the virus recovery is very low and mass balance between input and output doesn’t exist. It is speculated that that the majority of virus remains bound to the column after elution and high salt wash and only comes out in CIP step with NaOH. Virus titer determination was not possible in this fraction because of very high pH. However, the very sharp UV280 peak can be partly attributed to the remaining viruses coming out of the column. The full chromatogram of DMAE and CHTII are shown in Figure 5.
The overall results suggest that the steps and treatments in HPV-16 L1 VLP purification process are highly effective for baculovirus removal with overall LRV=17.
H2O2, UV-C, pasteurization, pH=4, pH=10 and Triton X-100 resulted in 5.58, 2.58, 5.87, 4.33, 0.95 and 3.62 LRV respectively. H2O2 potential in viral clearance have not been fully used in academic and industrial sections and few papers have considered its use for cell therapy products [13]. The high clearance is in agreement with other studies demonstrating large LRVs for other viruses [14]. Its high LRV suggests that it may be substituted for the virus filters which are costly and cannot be used for products which their molecules of interest are large and are filtered out by virus filters. Ease of use and removal H2O2 after treatment are advantages to this method. Furthermore, Triton X-100 having a rather high LRV is good news for the recombinant Influenza vaccine produced by BEVS, FluBlok, which uses Triton for protein extraction in the purification process [15].
High LRV by pasteurization is in agreement with the fact that many baculovirus proteins are inactivated by heat [16].
Conclusion
In addition to incorporation of the inactivation methods investigated in this study, into the purification process of biopharmaceuticals for viral clearance, these methods can also be used to ensure cleaning of production lines using BEVS technology especially for campaign production.
In conclusion, baculovirus can be removed or inactivated with numerous ways which the manufacturer can choose based on its product characteristics. And at last, it is worth mentioning that although these data provide a good reference to start with, it may not be generalized for the conditions employed during production of different products. Thus actual conditions of the production for a product to be marketed should be evaluated on a case-by-case basis for baculovirus removal or inactivation.
Acknowledgements
This study has been financially supported by Nivad Pharmed Salamat Company. The authors express their gratitude to Livogen company for providing an opportunity to use their Good Laboratory Practice (GLP) facilities for the development of qPCR assay and performing virus spiking studies and also to Noyan Biopharma company for providing spike starting materials.
Conflict of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper. Inclusion or exclusion of particular brands of instruments or reagents do not constitute an endorsement of these brands or vendors.
References
- Geigert J, Geigert J. The challenge of CMC regulatory compliance for biopharmaceuticals. New York, NY: Kluwer Academic/Plenum Publishers. (2004).
- Guideline IH. Viral safety evaluation of biotechnology products derived from cell lines of human or animal origin Q5A (R1). Current Step. (1997).
- Hefferon KL. Baculovirus late expression factors. J Mol Microbiol Biotechnol. 7, 89-101 (2004).
[Crossref] [Google Scholar] [PubMed]
- Cameron R, Smith K. Virus clearance methods applied in bioprocessing operations: An overview of selected inactivation and removal methods. Pharm Bioprocess. 2, 75-83 (2014).
- Debrus S, Martin MT, Stephen RJ, et al. Vaccine against HPV16 and HPV18 and at least another HPV type selected from HPV 31, 45 or 52. US Pat. (2010).
- Ghasemi A, Bozorg A, Rahmati F, et al. Comprehensive study on Wave bioreactor system to scale up the cultivation of and recombinant protein expression in baculovirus-infected insect cells. Biochem Eng. 143, 121-130 (2019).
- Mirhassani R, Seyedjafari E. Kinetics of infected insect cell osmolysis and enhanced protein release using a modified disruption method. Bioprocess Biosyst Eng. 39, 1729-1735 (2016).
[Crossref] [Google Scholar] [PubMed]
- Roldao A, Oliveira R, Carrondo MJ, Alve et al. Error assessment in recombinant baculovirus titration: Evaluation of different methods. J Virol Methods. 159, 69-80 (2009).
[Crossref] [Google Scholar] [PubMed]
- Litamoi J. Quality control testing of contagious bovine pleuropneumonia live attenuated vaccine: Standard operating procedures. FAO. (1996).
- World Health Organization. Global market study: HPV vaccines. World Health Organization. (2019).
- Brorson K, Lute S, Haque M, et al. A consensus rating method for small virus-retentive filters. II. Method evaluation. PDA J Pharm Sci Technol. 62, 334-343 (2008).
[Google Scholar] [PubMed]
- Carver DH, Seto DS. Viral inactivation by disulfide bond reducing agents. Virol J. 2, 1482-1484 (1968).
[Crossref] [Google Scholar] [PubMed]
- DePaula CA, Mahony DJ, Sunwoo MH, et al. A new process for viral clearance of cortical bone allografts and the effects on the mechanical properties and osteoinductivity. InPrograms and abstracts of the third annual world congress on tissue banking and 26th annual American association of tissue banks. American Association of Tissue Banks, Boston. (2002).
- Pinto AK, Richner JM, Poore, et al. A hydrogen peroxide-inactivated virus vaccine elicits humoral and cellular immunity and protects against lethal West Nile virus infection in aged mice. J Virol. 87, 1926-1936 (2013).
[Crossref] [Google Scholar] [PubMed]
- Bernardino TC. Expression of Rabies Virus Glycoprotein G (RVGP) and Matrix protein (M) using recombinant baculovirus. University of Sao Paulo, Brazil. (2019).
- Rohrmann GF. Baculovirus molecular biology. (2008).
[Google Scholar] [PubMed]