Editorial - Research on Chronic Diseases (2017) Volume 1, Issue 2
Coenzyme-A dependent catalysis: An overview of thiolase superfamily enzymes and drug discovery
- *Corresponding Author:
- Rajesh K. Harijan
Albert Einstein College of Medicine
New York, USA
E-mail: rajesh.harijan@einstein.yu.edu or rajesh.phd01@gmail.com
Abstract
Keywords
thiolases, lipid metabolism, infectious diseases, drug discovery
Abbreviations
CT: Cytosolic Thiolase; T1: Mitochondrial T1- thiolase; T2: Mitochondrial T2- thiolase; SCP2: Peroxisomal SCP2- thiolase; AB: Peroxisomal ABthiolase, TFE: Mitochondrial Trifunctional Enzyme Thiolase.
Editorial
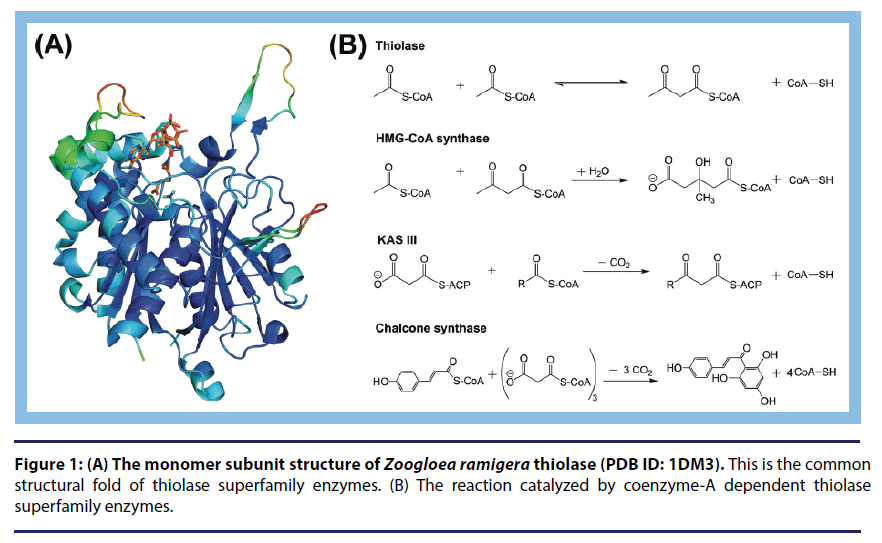
This is my first editorial article after becoming the board member of Research on Chronic Diseases. In this article, I would like to highlight the importance of coenzyme-A (CoA) dependent enzymes, particularly thiolases, for drug discovery research. CoA dependent enzymes are essential in various metabolic pathways. Approximately 4% of enzymes in the cells are CoA dependent and they are often involved in lipid/fatty acid metabolism (Figure 1) [1]. Lipid molecules are essential for the viability of all forms of life. These molecules are not only important constituents of the cell membrane but also used for producing energy in cells [2,3]. Thiolase superfamily is an important class of enzymes that belong to CoA dependent enzyme group. These enzymes are involved in fatty acid biosynthesis by carbon-carbon bond formation via a thioester-dependent Claisen-condensation reaction. They also play an important role in the degradation of fatty acids in the β-oxidation pathway [4]. 3-Ketoacyl-ACP synthases (KASs) are thiolase superfamily enzymes that are involved in fatty acid biosynthesis. There are two types of fatty acid synthesis (FAS) pathways known, namely FAS-I and FAS-II. FAS-I pathway is catalyzed by a large multi-domain protein complex, whereas FAS-II pathway is catalyzed by monofunctional enzymes [5]. In prokaryotes, fatty acids are synthesized by FAS-II pathway [6]. In eukaryotic cells, fatty acid synthesis is compartmentalized in the cytosol and mitochondria (and chloroplast in plants). The eukaryotic cytosol has the FAS-I fatty acid synthesis system, whereas mitochondria have the FAS-II [7]. Fatty acid synthesizing enzymes are mostly acyl carrier protein (ACP) dependent.
Thiolases are another set of enzymes that catalyze degradation of fatty acids in the β-oxidation pathway, which is the reversal of the fatty acid synthetic pathway. The main difference between two pathways is that the fatty acid synthesis is ACP dependent, whereas the fatty acid degradation is CoA dependent. Also, the fatty acid synthesis is NADP dependent, whereas β-oxidation is NAD dependent [8]. There are four different steps including dehydrogenation, hydration, oxidation and thiolytic cleavage that are involved in the β-oxidation pathway. The first reaction is catalysed by acyl-CoA dehydrogenase (mitochondrial) or acyl-CoA oxidase (peroxisomal) in which the trans-2 form of acyl-CoA molecules is generated. The second involves the hydration of trans-2 acyl- CoA into 3-hydroxyacyl-CoA. This step is catalyzed by hexameric enoyl-CoA hydratase [9]. The mitochondrial hydration is also done by the trifunctional enzyme (TFE, also producing L-3-hydroxyacyl-CoA), which also catalyzes the third and fourth steps of β-oxidation [10]. In peroxisome, the second and third steps are catalyzed by MFE1 and MFE2 [8]. MFE1 produces the L-3-hydroxyacyl-CoA intermediate, whereas for MFE2, the intermediate is R-3- hydroxyacyl-CoA. In mitochondria, the third step is catalyzed by a monofunctional 3-hydroxyacyl-CoA dehydrogenase. In this reaction, the L-3-hydroxyacyl-CoA molecule is converted into 3-ketoacyl-CoA, which undergoes thiolytic cleavage in the last (fourth) step catalyzed by thiolase enzyme.
Six different thiolases (CT, T1, T2, AB, SCP2 and TFE) have been discovered in the cytosol, mitochondria and peroxisomes of human cells. They occur as dimers or tetramers and have different enzyme kinetic properties and substrate specificities (4, 11). Enzymes of the thiolase superfamily share a conserved characteristic structure referred as the thiolase fold (βαβαβαββ; Figure 1) [11-13]. The first thiolase structure was discovered in the Wierenga laboratory in 1994 [12]. The polypeptide chain of thiolases consists of approximately 400 amino acid residues folded into three distinct domains: an N-domain, a loop region, and a C-domain. The residues important for CoA binding belong mostly to the loop domain. The catalytic residues reside in the loops of the core domains. Thiolases have four sequence fingerprints in their catalytic loops, including CxS-, NEAF-, GHP-, and CxG-motifs (4). CxS-cysteine is the nucleophilic cysteine of Nβ3-Nα3. The nucleophilic cysteine gets acylated during the reaction. The NEAF-motif belongs to the Cβ2-Cα2 loop and the asparagine side chain of this motif forms the oxyanion hole-1 (OAH1), which is involved in the stabilization of the negative charge on the thioester oxygen atom formed during catalysis. In SCP2- and TFE-thiolases, this sequence fingerprint is HDCF and HEAF, respectively. The histidine of GHP-loop (in the Cβ3-Cα3 loop) is also a hydrogen bond donor to OAH1 and is important for activating the nucleophilic cysteine. The CxG-motif belongs to the Cβ4- Cβ5 loop and the cysteine of this loop acts as acid/base [4,14].
Several other enzymes have been found to be structurally related to thiolases described above and consequently a thiolase superfamily has been formed, which includes not only the degradative and biosynthetic thiolases, but also other enzymes like the chalcone synthases (CHSs) and polyketide synthases (PKSs), as well as the HMG-CoA synthases [15]. To the best of my knowledge the enzymes of these three subclasses occur always as dimers.
The catalytic diversity of these enzymes is of the great research interest. The crucial metabolic importance of these enzymes in lipid metabolism makes them important for drug discovery research. Fatty acid molecules are essential for the viability of all forms of life. Consequently, these enzymes are also important from a pharmaceutical point of view to target infectious microorganisms. There are very few inhibitors developed for thiolase superfamily enzymes; for example, platensimysin [16], platencin, thiolactomycin [17] and cerulenin [18] were developed for KAS enzymes. Another inhibitor known as isoniazid, used in the treatment of tuberculosis, is an inhibitor of Mycobacterium tuberculosis KAS. The HMGCoA synthase is also a target for the development of cholesterol lowering drugs [19,20]. Thiolase enzymes have not been explored enough for drug discovery. These enzymes are found in all organisms including prokaryotes and eukaryotes. Remarkably, although thiolases have the same structural fold, but the sequences and substrate specificities are very different. The thiolase distribution in pathogenic bacteria and parasites is very different; for example, MTB and MSM have 8 and 13 thiolases respectively, whereas protozoa parasites have 1-2 thiolases [14].
Recently, it has been reported that T2-thiolase is responsible for metabolic alterations in cancer cells. Genetic knockdown studies of T2-thiolase in H1299 lung cancer cells show reduction in cell proliferation under hypoxia environment [21]. Major research work towards the knockout and enzyme chemistry of thiolases would be essential for drug discovery against these enzymes.
References
- Daugherty M.Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J. Biol. Chem. 277(24), 21431-21439 (2002).
- Yeagle PL. Lipid regulation of cell membrane structure and function. FASEB. J. 3(7), 1833-1842 (1989).
- Edidin M. Lipids on the frontier: A century of cell-membrane bilayers. Nat. Rev. Mol. Cell. Biol. 4(5), 414-418 (2003).
- Haapalainen AM, Merilainen G, Wierenga RK. The thiolase superfamily: Condensing enzymes with diverse reaction specificities. Trends. Biochem. Sci. 31(1), 64-71 (2006).
- Chan DI, Vogel HJ. Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem. J. 430(1), 1-19 (2010).
- Hiltunen JK, Chen Z, Haapalainen AM et al. Mitochondrial fatty acid synthesis--an adopted set of enzymes making a pathway of major importance for the cellular metabolism. Prog. Lipid. Res. 49(1), 27-45 (2010).
- Heath RJ, Rock CO. The claisen condensation in biology. Nat. Prod. Rep. 19(5), 581-596 (2002).
- Bhaumik P, Koski MK, Glumoff T et al. Structural biology of the thioester-dependent degradation and synthesis of fatty acids. Curr. Opin. Struct. Biol. 15(6), 621-628 (2005).
- Kim JJ, Battaile KP. Burning fat: The structural basis of fatty acid beta-oxidation. Curr. Opin. Struct. Biol. 12(6), 721-728 (2002).
- Ishikawa M, Tsuchiya D. Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex. EMBO. J. 23(14), 2745-2754 (2004).
- Fukao T. In Wiley Encyclopedia of Molecular Medicine, eds; Burchell J & Taylor-Papadimitriou J (John Wiley & Sons, Inc, pp: 3125-3128 (2002).
- Mathieu M.The 2.8 A crystal structure of peroxisomal 3-ketoacyl-CoA thiolase of saccharomyces cerevisiae: A five-layered alpha beta alpha beta alpha structure constructed from two core domains of identical topology. Structure. 2(9), 797-808 (1994).
- Mathieu M. The 1.8 A crystal structure of the dimeric peroxisomal 3-ketoacyl-CoA thiolase of saccharomyces cerevisiae: Implications for substrate binding and reaction mechanism. J. Mol. Biol. 273(3), 714-728 (1997).
- Anbazhagan P. Phylogenetic relationships and classification of thiolases and thiolase like proteins of mycobacterium tuberculosis and mycobacterium smegmatis. Tuberculosis. (Edinb). 94(4), 405-412 (2014).
- Jiang C, Kim SY, Suh DY. Divergent evolution of the thiolase superfamily and chalcone synthase family. Mol. Phylogenet. Evol. 49(3), 691-701 (2008).
- Wang J. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 441(7091), 358-361 (2006).
- Oishi H.Thiolactomycin, a new antibiotic. I. taxonomy of the producing organism, fermentation and biological properties. J. Antibiot. (Tokyo). 35(4), 391-395 (1982).
- Sato Y, Nomura S, Kamio Y et al. Studies on cerulenin, 3. isolation and physico-chemical properties of cerulenin. J. Antibiot. (Tokyo). 20(6), 344-348 (1967).
- Bahnson BJ. An atomic-resolution mechanism of 3-hydroxy-3-methylglutaryl-CoA synthase. Proc. Natl. Acad. Sci U S A 101(47), 16399-16400 (2004).
- Theisen MJ. 3-hydroxy-3-methylglutaryl-CoA synthase intermediate complex observed in "real-time". Proc. Natl. Acad. Sci. U S A. 101(47), 16442-16447 (2004).
- Fan J. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol. Cell. 53(4), 534-548 (2014).