Research Article - Clinical Practice (2018) Volume 15, Issue 2
Prevalence of aminoglycoside resistance genes in Pseudomonas aeruginosa isolated from a tertiary care hospital in Makkah, KSA
- Corresponding Author:
- Omar B Ahmed
Department of Environmental and Health Research
The Custodian of the Two Holy Mosques Institute for Hajj and Umrah
Umm Al-Qura University, Makkah, Saudi Arabia
E-mail: abuaglah1@hotmail.com
Abstract
Aminoglycosides are the most frequently prescribed antimicrobial agents in Saudi Arabia; they are routinely used for the treatment of gram-negative bacillary infections. The aim of this study was to detect the resistance patterns against different aminoglycoside antibiotics and the prevalence of the genes encoding for resistance in Pseudomonas aeruginosa isolates from Hera General Hospital, Makkah, KSA. All isolates that were resistant to one or more aminoglycoside antibiotic were subjected to antibiotic susceptibility test and PCR analysis to detect the presence of the resistance genes: aac (6’)-Ib, aac(3)-Ia, aac(3)-II , ant(2’’)-Ia, rmtB, rmtC, armA, rmtA, rmtD, rmtE, and npmA. The results showed that 46.1% of the isolates were resistant to one or more aminoglycoside antibiotics, but only 43.3% of these aminoglycoside-resistant isolates harbored resistance genes. In addition, 43% of the isolates were resistant to ciprofloxacin, amikacin, and ceftazidime. The resistance genes most frequently observed in these isolates were rmtB (7.6%) followed by aac (6’)-Ib (6.1%), rmtC (4.6%), and armA (1.5%). Taken together, these results indicate that the aminoglycoside-resistance genes are highly prevalent and could easily spread among P. aeruginosa strains. Coordinated efforts and further research works are needed to control antibiotic resistance to aminoglycosides before to be a threatening crisis.
Keywords
aminoglycoside, resistance genes, P. aeruginosa, phosphotransferase, modifying enzyme
Introduction
Aminoglycosides are highly potent, broadspectrum antibiotics with many desirable properties used for the treatment of lifethreatening infections [1]. Aminoglycosides are the most frequently prescribed antimicrobial agents in Saudi Arabia; they have been definitively established for the treatment of gram-negative bacillary infections. Aminoglycosides include many different agents such as gentamicin, tobramycin, amikacin, streptomycin, neomycin, and paromomycin; gentamicin, tobramycin, and amikacin are the most frequently prescribed. Aminoglycosides act primarily by binding to the aminoacyl site of the 16S ribosomal RNA within the 30S ribosomal subunit, leading to misreading of the genetic code and inhibition of translocation [2,3]. Aminoglycosides initially penetrate the organism by disrupting the magnesium and calcium bridges between lipopolysaccharide moieties. They are then transported across the cytoplasmic membrane in an energy-dependent manner. This step can be inhibited in vitro by divalent cations, increased osmolality, acidic pH, and an anaerobic environment [3]. Aminoglycoside resistance can occur through the acquisition or upregulation of genes that encode inactivating enzymes or efflux systems. Bacterial production of inactivating enzymes is the most common aminoglycoside resistance mechanism in gram-negative organisms [3]; resistance is due to the inactivation of aminoglycoside modifying enzymes (AMEs; aminoglycoside phosphotransferases, acetyl-transferases, and nucleotidyl-transferases) by the products of genes in plasmids or transposons. For example, the enzyme encoded by the rmtA gene has been associated with high-level resistance against all parenteral aminoglycosides currently in use [4,5]. The emergence of resistant strains has somewhat reduced the potential of aminoglycosides in empiric therapies [6]. Multidrug-resistant Pseudomonas aeruginosa have been emerging worldwide. The most common aminoglycosidemodifying enzyme gene types in P. aeruginosa are aac(6’)-I, aac(6’)-II, ant(2’’)-I, and aph(3’)- [7,8] and their substrates are the most important antipseudomonal aminoglycosides [9]. To date, no studies have been conducted in Saudi Arabia to determine which genes confer resistance to aminoglycosides in P. aeruginosa. This study aimed to detect the resistance patterns against different aminoglycoside antibiotics and the prevalence of the genes encoding for resistance in P. aeruginosa isolates from Makkah hospitals.
Material and Methods
This study involved the use of previously collected P. aeruginosa isolates by the authors and published [10] from Hera General Hospital (HGH)-Makkah; 263 beds). A total of 65 non-duplicated P. aeruginosa clinical isolates were identified from HGH. The frequency of isolates according to the wards as follows; male ward (n=13) female ward (n=10), Intensive care unit (n=11), surgery ward (n=10), obstetrics and Gynaecology (n=7), newborn intensive care unit (n=4), nursery (n=5) and pediatrics (n=3). The frequencies of isolates were: abscess (n=20), axillary (n=11), high vaginal swabs (n=10), pleural fluid (n=5), pus swab (n=8) and sputum sample (n=9). The study was approved by Ethics Committee in The Custodian of the Two Holy Mosques Institute for Hajj and Umrah, Umm Al-Qura University and a written informed consent has been taken from the subjects participating in the study.
Determination of antibiotic susceptibility for P. aeruginosa isolates
A standardized inoculum (1 × 108 CFU/mL) of all isolates was inoculated on the surface of a large (150 mm diameter) Mueller-Hinton agar plate at 35°C. The antimicrobial susceptibility of all clinical isolates was examined using the disc diffusion method with various antibiotics (TABLE 1). The tested antibiotics were ceftazidime (CAZ) (30 μg), cefotaxime (CTX) (30 μg), Ciprofloxacin (CIP) (10 μg) amikacin (AK) (30 μg), piperacillin/tazobactum (TZP) (100/10) (CRO) (30 μg), cefepime (FEB) (30), imipenam (IPM) (10), colistin (Cl) (10 μg), gentamycin (GN)(10 μg) and tobramycin (TOB) (10 μg).
| Antibiotic | Abbreviation | Concentration (µg) |
|---|---|---|
| Ceftazidime | CAZ | 30 |
| Cefotaxime | CTX | 30 |
| Ciprofloxacin | CIP | 10 |
| Amikacin | AK | 30 |
| Cefepime | FEP | 30 |
| Piperacillin/Tazobactam | TZP | 100/10 |
| Imipenem | IPM | 10 |
| Colistin | CL | 10 |
| Gentamicin | GN | 10 |
| Tobramycin | TOB | 10 |
Table 1: Antibiotics used for susceptibility testing.
Antibiotic disks were placed on the inoculated agar surface. Plates were incubated for 16-24 h at 35°C. The zones of growth inhibition around each of the antibiotic disks were measured to the nearest millimeter. The diameter of the zone is related to the susceptibility of the isolate and to the diffusion rate of the drug through the agar medium. The zone diameters of each drug are interpreted using the using Clinical and Laboratory Standards Institute (CLSI) method [11]. CLSI provides for three categories of identification: susceptible, intermediate and resistant. Susceptible defines a level of antimicrobial activity associated with a high likelihood of therapeutic success. Intermediate defines a level of antimicrobial agent activity associated with uncertain therapeutic effect. Resistant defines a level of antimicrobial activity associated with a high likelihood of therapeutic failure [11].
DNA extraction
DNA was prepared by guanidinium thiocyanate extraction, as previously described [12]. Two bacterial colonies (3 mm in diameter) were collected from the nutrient agar plates and dispersed in 100 mL of 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. The cells were lysed with 500 mL GES reagent [5 M guanidinium thiocyanate (Sigma, city,state (abbrev), country), 0.1 M EDTA, and 0.5% (w/v) sarcosyl (Sigma)]. Following the addition of 250 mL 7.5 M ammonium acetate, the suspension was kept on ice for 10 minutes. For deproteination, 500 mL chloroform:isoamyl alcohol (24:1) was added and the mixture was centrifuged at 13,000 g for 10 minutes. The DNA was precipitated from the upper phase with 100% ethanol at -20°C for 1 hour. The extracted DNA was used as a template for PCR amplification.
PCR analysis
All isolates that were resistant to one or more aminoglycoside antibiotic were subjected to PCR analysis to detect the presence of the following resistance genes: aac(6’)-Ib, aac(3)- Ia, aac(3)-II , ant(2’’)-Ia, rmtB, rmtC, armA, rmtA, rmtD, rmtE, and npmA (TABLE 2). PCR was performed in a final volume of 25 μL. The primers used for PCR amplification are listed in TABLE 2. Each reaction contained 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 0.2 mM of each deoxynucleoside triphosphate, 1.5 mM MgCl2, 1.5 μL of each primer, 1.25 U of Taq DNA polymerase, and 2 μL of template DNA. Two multiplex reactions were performed. The first reaction included the aac(6’)-Ib, aac(3)-II genes using the following conditions: pre-denaturation at 94°C for 4 minutes; 35 amplification cycles of 94°C for 1 minute, 55°C for 1 minute, and 72°C for 1.5 minutes; and a final extension step of 72°C for 5 minutes. The second reaction included the armA, rmtB, rmtC, and rmtD genes using the following conditions: pre-denaturation at 94°C for 4 minutes; 35 amplification cycles of 94°C for 1 minute, 50°C for 1 minute, and 72°C for 1.5 minutes; and a final extension step of 72°C for 5 minutes. Amplified PCR products were detected by agarose gel electrophoresis. A DNA marker (Promega, USA) was run with each gel and the genotype was determined by the size of the amplified product.
| No. | Gene | Primer sequence | Product size (bp) | Reference |
|---|---|---|---|---|
| 1 | aac(6')-lb-F | TTG CGA TGC TCT ATG AGT GGC TA | 472 | aac(6’)-Ib [7,8] |
| aac(6')-lb-R | CTC GAA TGC CTG GCG TGT TT | |||
| 2 | aac(3)-II-F | ATATCGCGATGCATACGCGG | 877 | aac(3)-II [7,8] |
| aac(3)-II-R | GACGGCCTCTAACCGGAAGG | |||
| 3 | rmtB-F | GCT TTC TGC GGG CGA TGT AA | 173 | rmtB [4,5] |
| rmtB-R | ATG CAA TGC CGC GCT CGT AT | |||
| 4 | rmtC-F | CGA AGA AGT AAC AGC CAA AG | 711 | rmtC [4,5] |
| rmtC-R | ATC CCA ACA TCT CTC CCA CT | |||
| 5 | armA-F | ATT CTG CCT ATC CTA ATT GG | 315 | armA [4,5] |
| armA-R | ACC TAT ACT TTA TCG TCG TC | |||
| 6 | rmtD-F | CGG CAC GCG ATT GGG AAG C | 401 | rmtD [4,5] |
| rmtD-R | CGG AAA CGA TGC GAC GAT | |||
| 7 | npmA-F | CTC AAA GGA ACA AAG ACG G | 774 | npmA [4,5] |
| npmA-R | GAA ACA TGG CCA GAA ACT C |
Table 2: Primers used in the study.
Statistical analysis
Statistical analysis was carried out using Statistical Package for Social Sciences (SPSS) software (version 21.0) for Windows (x2-test). A p value of =>0.05 was considered significant.
Results
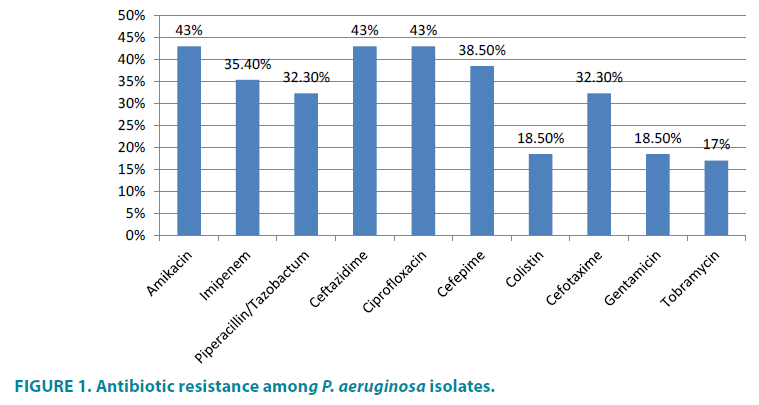
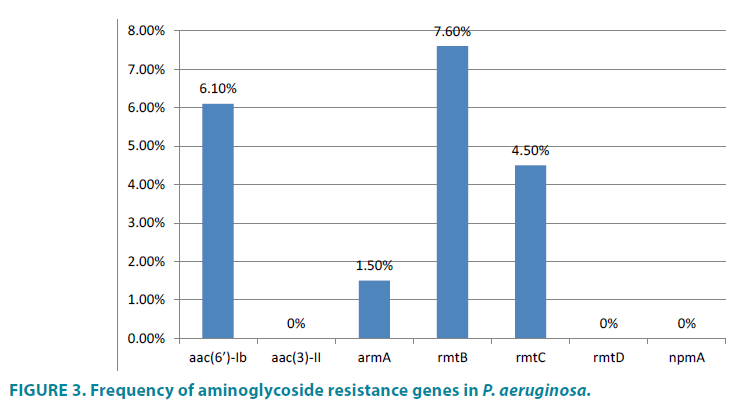
The resistance profiles of the P. aeruginosa isolates showed that 46.10% (30/65) were resistant to one or more aminoglycoside antibiotics and that only 43.3% (13/30) of the aminoglycoside-resistant isolates harbored resistance genes; none of the susceptible isolates harbored the tested resistance genes. In addition, all of the isolates contained only a single aminoglycoside modifying gene - i.e. none of the isolates co-harbored more than one resistance gene. The results of the present study showed that 43% of the isolates (28/65) were resistant to ciprofloxacin, amikacin, and ceftazidime while 35.4% (23/65) were resistant to imipenem and 32.3% (21/65) were resistant to piperacillin-tazobactam (FIGURES 1 and 4). In addition, 18.5% (12/65) and 17% (11/65) of the isolates exhibited resistance to gentamicin and tobramycin, respectively (FIGURE 1). The resistance genes observed most frequently in these isolates were rmtB (7.6%, 5/65), aac(6’)-Ib, (6.1%, 4/65), rmtC (4.6%, 3/65), and armA (1.5%, 1/65) (FIGURES 2 and 3). Moreover, none of the susceptible isolates harbored these resistance genes. The difference in the distribution of the resistance genes was statistically significant between the various aminoglycosides antibiotic resistance profiles (P value <0.05).
Figure 2: Aminoglycoside resistance genes detected by PCR and 2% agarose gel electrophoresis. Lane 1: rmtC positive (711 bp), Lane 2: rmtB positive (173 bp), Lane 3: armA positive (315 bp), Lane M: 100-bp DNA ladder.
Discussion
Aminoglycosides are broad-spectrum antibiotics of high potency that have been traditionally used for the treatment of serious gram-negative bacteria such as Pseudomonas infections [13]. Aminoglycosides act by inhibiting protein synthesis via binding to the 16S rRNA and by disrupting bacterial cell membrane integrity [14]. P. aeruginosa is one of the most prevalent hospital acquired pathogens associated with higher mortality rates and antibiotic costs. In the present study, a total of 65 non-duplicated P. aeruginosa clinical isolates were identified in a tertiary hospital in Makkah. Treatment of P. aeruginosa is complicated by its ability to develop resistance to multiple classes of antibacterial agents, even during the course of infection treatment. In the present study, the antimicrobial susceptibility of P. aeruginosa strains isolated from HGH was tested. The resistance profiles showed that 46.1% were resistant to one or more antibiotics and 43% were resistant to amikacin, ciprofloxacin, and ceftazidime, as well as imipenem (35.4%), and piperacillin-tazobactam (32.3%). In addition, 18.5% and 17.0% of the isolates were resistant to the aminoglycosides gentamicin and tobramycin, respectively. In this study, P. aeruginosa exhibited 43% resistance against ciprofloxacin, which is in agreement with other studies [15-18]. Resistance to fluoroquinolones is most likely due to mutation as the result of selective pressure created by the use of fluoroquinolones [19].
Carbapenems are often used as a lastresort treatment for Pseudomonas infections. Resistance to imipenems was shown to be 35.4%. This result is in accordance with a other reports [20,21].
The detected resistance rates against the aminoglycosides amikacin, gentamicin, and tobramycin were 43%, 18.5%, and 17% respectively. The overall incidence of aminoglycoside resistance found in our study was lower than previously reported in other countries worldwide [22-24]. However, a number of studies have reported a high level of aminoglycoside resistance (amikacin and gentamycin) in pseudomonal infection cases [25]. A high frequency of P. aeruginosa isolates resistant to tobramycin and amikacin has also been reported in other health institutions [26]. Another study has reported 27% resistance to gentamicin, 19% to amikacin, and 23% to tobramycin [27]; while 24.2% resistance to gentamicin and 16.7% resistance to amikacin have been reported in Spain [28].
In the present study, 46.1% of the P. aeruginosa isolates were resistant to one or more aminoglycosides antibiotics, while only 43.3% of these aminoglycoside-resistant isolates harbored resistance genes. Statistically, there is a significant difference in the frequency of resistance genes across the aminoglycosides resistance profiles (P value=<0.05). These results highlight the importance of aminoglycosidemodification- related mechanisms in aminoglycoside resistance in P. aeruginosa. The resistance genes most frequently observed in these isolates were rmtB (7.6%) and aac(6’)- Ib (6.1%). Previous studies have demonstrated the existence of the rmtA and rmtB genes in P. aeruginosa isolates [29,30]. The results of this study are similar to other studies conducted in different countries [31-33]. None of the susceptible isolates harbored resistance genes, in agreement with several other studies conducted around the world [34]. The most common aminoglycoside resistance determinants found in P. aeruginosa are aac(6´)-II and ant(2´´)-I in Europe; aph(3′)-VI, ant(2´´)-I, and aac(6´)-I in Korea; and aac(6´)-31/aadA1 and aadA2 in Mexico and Brazil [35,36]. This difference in the distribution of aminoglycoside genes may be attributed to differences in aminoglycoside prescription patterns, selection of bacterial populations, or geographical differences in the occurrence of aminoglycoside resistance genes [9]. In conclusion, these aminoglycosideresistance genes are highly prevalent and could easily spread among P. aeruginosa strains [34]. Coordinated efforts and new research are needed to control antibiotic resistance to aminoglycosides before to be a threatening crisis.
Acknowledgements
The authors are grateful to The Custodian of the Two Holy Mosques Institute for Hajj and Umrah, Umm Al-Qura University, Makkah, Saudi Arabia., for supporting this study.
References
- Gilbert DN. Aminoglycosides. Principles and practice of infectious diseases. 4th ed. New York, NY: Churchill Livingstone pp. 279-306 (1995).
- Shakil S, Khan R, Zarrilli R, Khan AU. Aminoglycosides versus bacteria–a description of the action, resistance mechanism, and nosocomial battleground. J. Biomed. Sci. 15, 5-14 (2008).
- Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. Aminoglycosides: activity and resistance. Antimicrob. Agents Chemother. 43, 727 (1999).
- Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45, 88-94 (2007).
- Kashfi M, Hashemi A, Eslami G, et al. The Prevalence of Aminoglycoside-modifying Enzyme Genes among Pseudomonas aeruginosa Strains Isolated From Burn Patients. Arch. Clin. Infect Dis. 12(1), e40896 (2017).
- Murray BE. New aspects of antimicrobial resistance and the resulting therapeutic dilemmas. J. Infect. Dis. 163, 1185-1194 (1991).
- Gemmell CG, Edwards DI, Fraise AP, et al. Guidelines for the prophylaxis and treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections in the UK. J. Antimicrob. Chemother. 57, 589 (2006).
- Yokoyama K, Doi Y, Yamane K, et al. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362, 1888 (2003).
- Vaziri F, Peerayeh SN, Nejad QB, Farhadian A. The prevalence of aminoglycoside-modifying enzyme genes (aac (6’)-I, aac (6’)-II, ant (2”)-I, aph (3’)-VI) in Pseudomonas aeruginosa. Clinics(Sao Paulo). 66, 1519-1522 (2011).
- Ahmed OB, Asghar AH. Antibiotic susceptibility pattern of Pseudomonas aeruginosa expressing blaGES and blaPER genes in two different hospitals. African J. Biotechnol. 16(21), 1197-1202 (2017).
- Clinical and Laboratory Standards Institute, Performance standards for antimicrobial susceptibility testing. Nineteenth informational supplement M100-S19, Wayne, PA, Clinical and Laboratory Standards Institute (2009).
- Pitcher D, Saunders N, Owen R. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8, 151-156 (1989).
- Hermann T. Aminoglycoside antibiotics: old drugs and new therapeutic approaches. Cell Mol. Life Sci. 64, 1841-1852 (2007).
- Shakil S, Khan R, Zarrilli R, Khan AU. Aminoglycosides versus bacteria–a description of the action, resistance mechanism, and nosocomial battleground. J. Biomed. Sci. 15, 5-14 (2008).
- Ali S Q, Zehra A, Naqvi BS, Shah S, Bushra R. Resistance Pattern of Ciprofloxacin Against Different Pathogens. Oman Med. J. 25(4), 294-298 (2010).
- Hu XH, Xu XM, Mi ZH, Fan YF, Feng WY. Relationship between drug resistance of Pseudomonas aeruginosa isolated from burn wounds and its mobile genetic elements. Zhonghua Shao Shang Za Zhi 25(2), 103-105 (2009).
- KM Mohanasoundaram. Antimicrobial resistance in pseudomonas aeruginosa. J. Clin. Diagn. Res. 5(3), 491-494 (2011).
- Marilee DO, Douglas NF, Robert ML, Rose J. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob. Agents Chemother. 48(12), 4606-4610 (2004).
- Sheng WH, Chen YC, Wang JT, et al. Emerging fluoroquinolone-resistance for common clinically important gram negative bacteria in Taiwan. Diagn. Microbiol. Infect. Dis. 43, 141-147 (2002).
- Marilee DO, Douglas NF, Robert ML, Rose J. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob. Agents Chemother. 48(12), 4606-4610 (2004).
- Brown PD, Izundu A. Antibiotic resistance in clinical isolates of Pseudomonas aeruginosa in Jamaica. Rev Panam Salud Publica. 16, 125-130 (2004).
- Poole K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49, 479-487 (2005).
- Kim JY, Park YJ, Kwon HJ, et al. Occurrence and mechanisms of amikacin resistance and its association with b-lactamases in Pseudomonas aeruginosa: a Korean nationwide study. J. Antimicrob. Chemother. 62, 479-483 (2008).
- Cavallo J, Hocquet D, Plesiat P, Fabre R, Roussel-Delvallez M. Susceptibility of Pseudomonas aeruginosa to antimicrobials: a 2004 French multicentre hospital study. J. Antimicrob. Chemother. 59, 1021-1024 (2007).
- Estahbanati H, Kashani P, Ghanaatpisheh F. Frequency of Pseudomonas aeruginosa serotypes in burn wound infections and their resistance to antibiotics. Burns. 28, 340-348 (2002).
- Brito A, Landaeta JM, Roldán Y, et al. Resistencia de Pseudomonas aeruginosa a la gentamicina, tobramicina amikacina en Venezuela. Bol. Soc. Ven. Microbiol. 20, 42-45 (2000).
- Teixeira B, Rodulfo H, Carreño N, et al. Aminoglycoside resistance genes in Pseudomonas aeruginosa isolates from Cumana, Venezuela. Rev. Inst. Med. Trop. 58, 13 (2016).
- Gamero M, García-Mayorgas A, Rodríguez F, Ibarra A, Casal M. Sensibilidad y resistencia de Pseudomonas aeruginosa a los antimicrobianos. Rev. Esp. Quimioter. 20, 230-233 (2007).
- Islam S, Oh H, Jalal S, et al. Chromosomal mechanisms of aminoglycoside resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Clin. Microbiol. Infect.15, 60-66 (2009).
- Shaw KJ, Rather PN, Hare RS, Miller GH. Molecular geneticsaminoglycoside resistance genes and familial relationship of the aminoglycoside modifying enzymes. Microbiol. Rev. 57, 138-163 (1993).
- Busch-Sorensen C, Sonmezoglu M, Frimodt-Moller N, et al. Aminoglycoside resistance mechanisms in Enterobacteriaceae and Pseudomonas spp. from two Danish hospitals: correlation with type of aminoglycoside used. APMIS. 104, 763-768 (1996).
- Over U, Gur D, Unal S, Miller GH. The changing nature of aminoglycoside resistance mechanisms and prevalence of newly recognized resistance mechanisms in Turkey. Clin. Microbiol. Infect. 7, 470-478 (2001).
- Phillips I, King A, Shannon K. Prevalence and mechanisms of aminoglycoside resistance. A ten-year study. Am. J. Med. 80, 48-55 (1986).
- Miller GH, Sabatelli FJ, Hare RS, et al. The most frequent aminoglycoside resistance mechanisms changes with time and geographic area: a reflection of aminoglycoside usage patterns. Clin. Infect Dis. 24, S46-62 (1997).
- Mendes RE, Castanheira M, Toleman MA, et al. Characterization of an integron carrying blaIMP-1 and a new aminoglycoside resistance gene, aac(6’)-31, and its dissemination among genetically unrelated clinical isolates in a Brazilian hospital. Antimicrob. Agents Chemother. 51, 2611-264 (2007).
- Sánchez-Martinez G, Garza-Ramos UJ, Reyna-Flores FL, et al. In169, a new class 1 integron that encoded bla(IMP-18) in a multidrug-resistant Pseudomonas aeruginosa isolate from Mexico. Arch. Med. Res. 41, 235-239 (2010).